
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,289,651 – 20,289,781 |
| Length | 130 |
| Max. P | 0.724545 |

| Location | 20,289,651 – 20,289,744 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 77.57 |
| Shannon entropy | 0.37540 |
| G+C content | 0.52724 |
| Mean single sequence MFE | -27.12 |
| Consensus MFE | -18.54 |
| Energy contribution | -19.26 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.643046 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 20289651 93 - 27905053 CACUUGAGUUCGUGGCGCUCCCAAAGAGUGGAGAUAUCCUUUCAUAGGAGCGAACAAAGCGAACGGGGAGAAAUCCACACGGCUGCCUUGCUG .......((((((..((((((....(((.(((....))).)))...))))))......))))))..(((....)))...((((......)))) ( -32.40, z-score = -2.04, R) >droSim1.chr3R 20130621 77 - 27517382 CACUUGAGUUCGCUGUGCUCCCAAAGAGUGGAGAUAUCCUUUCAGAGGAGCGAACAAAGCGAACGGCUG-----CCUUGCUG----------- .......(((((((.((((((....(((.(((....))).)))...)))))).....)))))))(((..-----....))).----------- ( -27.70, z-score = -2.22, R) >droSec1.super_0 20626052 77 - 21120651 CACUUGAGUUCGCUGUGCUCCCAAAGAGUGGAGAUAUCCUUUCAGAGGAGCGAACAAAGCGAAUGGCUG-----CCUUACUG----------- .......(((((((.((((((....(((.(((....))).)))...)))))).....))))))).....-----........----------- ( -22.90, z-score = -1.24, R) >droYak2.chr3R 2618802 93 + 28832112 CACUCGAGUUCGCUGUGCUCCAAACGAGUGCAGAUAUCCUUUCAGAGGAGCGAACAAAGCCAACGAGGAGAAAUCCACACUGCAUCCUUGCUG .......((((((((..(((.....)))..))....(((((...)))))))))))...........(((....))).....(((....))).. ( -28.00, z-score = -1.82, R) >droEre2.scaffold_4820 2700341 92 + 10470090 CACUUGAGUUCUCUGUGCUCCAAACGAGUGCAGGUAUCCUUUCAGAGGAGCG-ACAAAGCGACCGGGGAGAAAUCCACACUGCCUCCUUGCUG ............(((..(((.....)))..)))((.(((((...)))))...-))..(((((..(((.((.........)).)))..))))). ( -24.60, z-score = 0.44, R) >consensus CACUUGAGUUCGCUGUGCUCCCAAAGAGUGGAGAUAUCCUUUCAGAGGAGCGAACAAAGCGAACGGGGAGAAAUCCACACUGC__CCUUGCUG .......(((((((.((((((....(((.(((....))).)))...)))))).....)))))))............................. (-18.54 = -19.26 + 0.72)


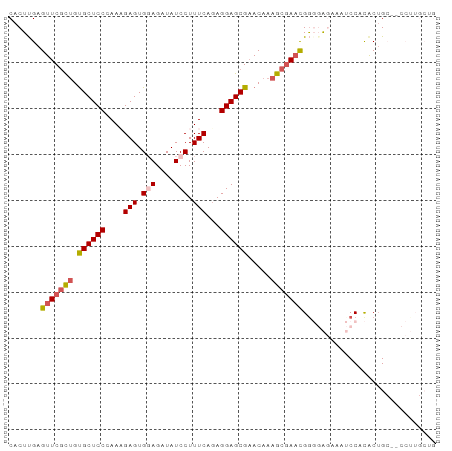
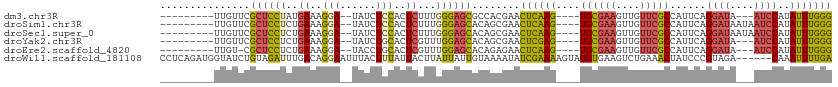
| Location | 20,289,687 – 20,289,781 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 73.68 |
| Shannon entropy | 0.45842 |
| G+C content | 0.43817 |
| Mean single sequence MFE | -25.13 |
| Consensus MFE | -15.93 |
| Energy contribution | -15.85 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.603639 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 20289687 94 + 27905053 ---------UUGUUCGCUCCUAUGAAAGGA--UAUCUCCACUCUUUGGGAGCGCCACGAACUCAAG----UGCGAAGUUGUUCGCCAUUCAGGAUA---AUCCAUAUUUGGG ---------((((.((((((((.((..(((--....)))..))..))))))))..)))).((((((----((((((....)))))......((...---..))..))))))) ( -28.90, z-score = -2.16, R) >droSim1.chr3R 20130641 97 + 27517382 ---------UUGUUCGCUCCUCUGAAAGGA--UAUCUCCACUCUUUGGGAGCACAGCGAACUCAAG----UGCGAAGUUGUUCGCCAUUCAGGAUAAUAAUCCAUAUUUGGG ---------..(((((((.((((.((((..--(......)..)))).))))...)))))))(((((----((((((....)))))......((((....))))..)))))). ( -28.70, z-score = -2.08, R) >droSec1.super_0 20626072 97 + 21120651 ---------UUGUUCGCUCCUCUGAAAGGA--UAUCUCCACUCUUUGGGAGCACAGCGAACUCAAG----UGCGAAGUUGUUCGCCAUUCAGGAUAAUAAUCCAUAUUUGGG ---------..(((((((.((((.((((..--(......)..)))).))))...)))))))(((((----((((((....)))))......((((....))))..)))))). ( -28.70, z-score = -2.08, R) >droYak2.chr3R 2618838 94 - 28832112 ---------UUGUUCGCUCCUCUGAAAGGA--UAUCUGCACUCGUUUGGAGCACAGCGAACUCGAG----UGCGAAGUUGUUCGCCAUUCAGGAUA---AUCCAUAUUUGGG ---------..(((((((.(((..((.(..--(......)..).))..)))...)))))))..(((----((((((....)))).)))))......---..(((....))). ( -25.50, z-score = -0.50, R) >droEre2.scaffold_4820 2700377 93 - 10470090 ---------UUGU-CGCUCCUCUGAAAGGA--UACCUGCACUCGUUUGGAGCACAGAGAACUCAAG----UGCGAAGUUGUUCGCCAUUCAGGAUA---AUCCAUAUUUGGG ---------((((-.(((((..(((.(((.--..)))....)))...)))))))))....((((((----((((((....)))))......((...---..))..))))))) ( -23.40, z-score = -0.25, R) >droWil1.scaffold_181108 297265 106 - 4707319 CCUCAGAUGGUAUCUGUAGAUUUGACAGGAAUUUACUUUAUUACUUAUUAUUGUAAAAUAUCGAAAAGUAUGUGAAGUCUGAAACUAUCCCGUAGA------CAAAUUUUGA .....((((((.((...((((((.((.....(((((..((........))..)))))((((......)))))).)))))))).)))))).......------.......... ( -15.60, z-score = 0.58, R) >consensus _________UUGUUCGCUCCUCUGAAAGGA__UAUCUCCACUCUUUGGGAGCACAGCGAACUCAAG____UGCGAAGUUGUUCGCCAUUCAGGAUA___AUCCAUAUUUGGG ...............((((((..((..(((......)))..))...))))))........((((((.....(((((....)))))......((((....))))...)))))) (-15.93 = -15.85 + -0.08)

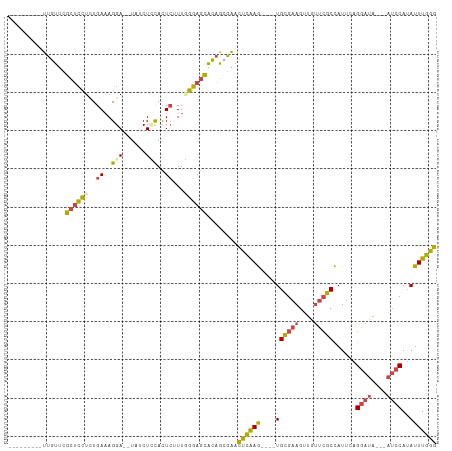
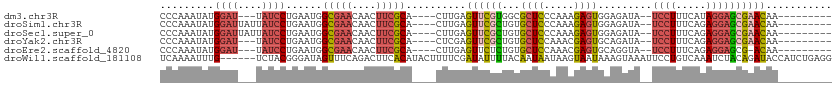
| Location | 20,289,687 – 20,289,781 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 73.68 |
| Shannon entropy | 0.45842 |
| G+C content | 0.43817 |
| Mean single sequence MFE | -24.08 |
| Consensus MFE | -14.64 |
| Energy contribution | -15.37 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.724545 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 20289687 94 - 27905053 CCCAAAUAUGGAU---UAUCCUGAAUGGCGAACAACUUCGCA----CUUGAGUUCGUGGCGCUCCCAAAGAGUGGAGAUA--UCCUUUCAUAGGAGCGAACAA--------- .(((....)))..---...(((((((.(((((....))))).----.....))))).))((((((....(((.(((....--))).)))...)))))).....--------- ( -25.50, z-score = -1.30, R) >droSim1.chr3R 20130641 97 - 27517382 CCCAAAUAUGGAUUAUUAUCCUGAAUGGCGAACAACUUCGCA----CUUGAGUUCGCUGUGCUCCCAAAGAGUGGAGAUA--UCCUUUCAGAGGAGCGAACAA--------- ..(((....((((....))))......(((((....))))).----.))).((((((....(((((.....).))))...--(((((...)))))))))))..--------- ( -26.80, z-score = -1.76, R) >droSec1.super_0 20626072 97 - 21120651 CCCAAAUAUGGAUUAUUAUCCUGAAUGGCGAACAACUUCGCA----CUUGAGUUCGCUGUGCUCCCAAAGAGUGGAGAUA--UCCUUUCAGAGGAGCGAACAA--------- ..(((....((((....))))......(((((....))))).----.))).((((((....(((((.....).))))...--(((((...)))))))))))..--------- ( -26.80, z-score = -1.76, R) >droYak2.chr3R 2618838 94 + 28832112 CCCAAAUAUGGAU---UAUCCUGAAUGGCGAACAACUUCGCA----CUCGAGUUCGCUGUGCUCCAAACGAGUGCAGAUA--UCCUUUCAGAGGAGCGAACAA--------- .(((....)))..---.....(((...(((((....))))).----.))).((((((((..(((.....)))..))....--(((((...)))))))))))..--------- ( -28.20, z-score = -2.36, R) >droEre2.scaffold_4820 2700377 93 + 10470090 CCCAAAUAUGGAU---UAUCCUGAAUGGCGAACAACUUCGCA----CUUGAGUUCUCUGUGCUCCAAACGAGUGCAGGUA--UCCUUUCAGAGGAGCG-ACAA--------- .(((....)))..---..((((.....(((((....))))).----.(((((...((((..(((.....)))..))))..--....)))))))))...-....--------- ( -23.20, z-score = -0.58, R) >droWil1.scaffold_181108 297265 106 + 4707319 UCAAAAUUUG------UCUACGGGAUAGUUUCAGACUUCACAUACUUUUCGAUAUUUUACAAUAAUAAGUAAUAAAGUAAAUUCCUGUCAAAUCUACAGAUACCAUCUGAGG .....(((((------...((((((.((((...)))).....((((((....(((((.........)))))..))))))...)))))))))))((.(((((...))))))). ( -14.00, z-score = 0.03, R) >consensus CCCAAAUAUGGAU___UAUCCUGAAUGGCGAACAACUUCGCA____CUUGAGUUCGCUGUGCUCCCAAAGAGUGGAGAUA__UCCUUUCAGAGGAGCGAACAA_________ .........((((....))))......(((((....)))))..........((((((...((((.....)))).........((((.....))))))))))........... (-14.64 = -15.37 + 0.73)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:40:45 2011