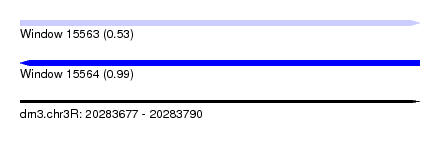
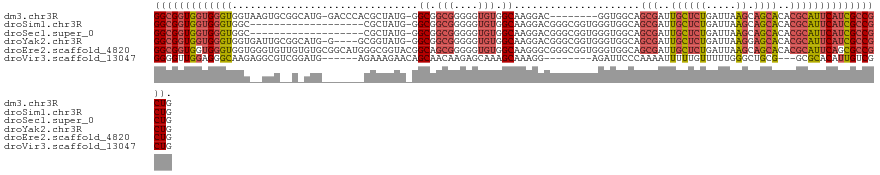
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,283,677 – 20,283,790 |
| Length | 113 |
| Max. P | 0.988781 |

| Location | 20,283,677 – 20,283,790 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 123 |
| Reading direction | forward |
| Mean pairwise identity | 68.86 |
| Shannon entropy | 0.57685 |
| G+C content | 0.64404 |
| Mean single sequence MFE | -28.21 |
| Consensus MFE | -13.47 |
| Energy contribution | -13.50 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.531538 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
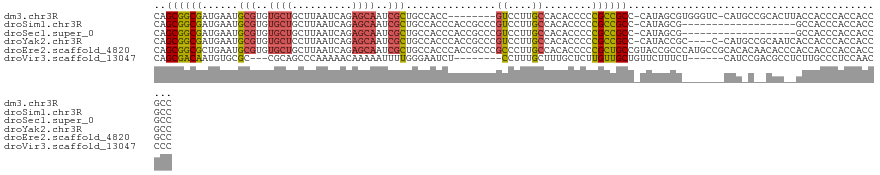
>dm3.chr3R 20283677 113 + 27905053 CAGCGGCGAUGAAUGCGUGUGCUGCUUAAUCAGAGCAAUCGCUGCCACC--------GUCCUUGCCACACCCCCGCCGCC-CAUAGCGUGGGUC-CAUGCCGCACUUACCACCCACCACCGCC ..(((((((((...(((.(((.(((((.....)))))..))))))...)--------)))........((((.(((....-....))).)))).-...))))).................... ( -33.00, z-score = -0.51, R) >droSim1.chr3R 20124792 103 + 27517382 CAGCGGCGAUGAAUGCGUGUGCUGCUUAAUCAGAGCAAUCGCUGCCACCCACCGCCCGUCCUUGCCACACCCCCGCCGCC-CAUAGCG-------------------GCCACCCACCACCGCC ..(((((((.((..(((.((((((......)))(((....)))......))))))...)).)))))........(((((.-....)))-------------------))...........)). ( -29.90, z-score = -1.37, R) >droSec1.super_0 20620270 103 + 21120651 CAGCGGCGAUGAAUGCGUGUGCUGCUUAAUCAGAGCAAUCGCUGCCACCCACCGCCCGUCCUUGCCACACCCCCGCCGCC-CAUAGCG-------------------GCCACCCACCACCGCC ..(((((((.((..(((.((((((......)))(((....)))......))))))...)).)))))........(((((.-....)))-------------------))...........)). ( -29.90, z-score = -1.37, R) >droYak2.chr3R 2612712 117 - 28832112 CAGCGGCGAUGAAUGCGUGUGCUCCUUAAUCAGAGCAAUCGCUGCCACCCACCGCCCGUCCUUGCCACACCCCCGCCGCC-CAUACCGC----C-CAUGCCGCAAUCACCACCCACCACCGCC ..(((((((((...(((..(((((........)))))..))).((........)).))))..............((.(..-....).))----.-...))))).................... ( -24.60, z-score = -0.67, R) >droEre2.scaffold_4820 2694459 123 - 10470090 CAGCGGCGCUGAAUGCGUGUGCUGCUUAAUCAGAGCAAUCGCUGCCACCCACCGCCCGCCCUUGCCACACCCCCGCUGCCGUACCGCCCAUGCCGCACACAACACCCACCACCCACCACCGCC ..((((((.((...(((.(((((((((.....)))))...((.((........((........)).........)).)).))))))).))))))))........................... ( -27.43, z-score = -0.09, R) >droVir3.scaffold_13047 3647464 106 + 19223366 CAGCGACAAUGUGCGC---CGCAGCCCAAAAACAAAAAUUUUGGGAAUCU--------CCUUUGCUUUGCUCUUGUUGCUGUUCUUUCU------CAUCCGACGCCUCUUGCCCUCCAACCCC (((((((((.(.(((.---.(((((((((((.......))))))).....--------...))))..))).))))))))))........------............................ ( -24.40, z-score = -2.02, R) >consensus CAGCGGCGAUGAAUGCGUGUGCUGCUUAAUCAGAGCAAUCGCUGCCACCCACCGCCCGUCCUUGCCACACCCCCGCCGCC_CAUAGCG_______CAU_C__CAC__ACCACCCACCACCGCC ..((((((......(((..((((..........))))..)))...............((....))........))))))............................................ (-13.47 = -13.50 + 0.03)
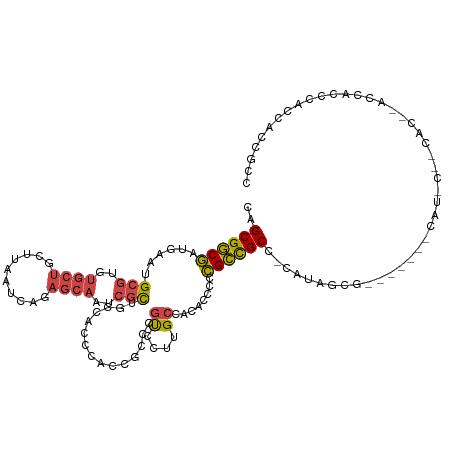
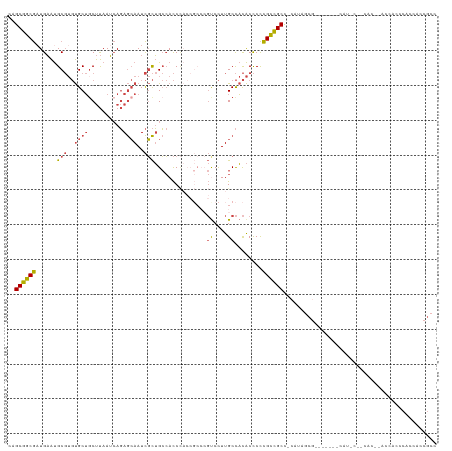
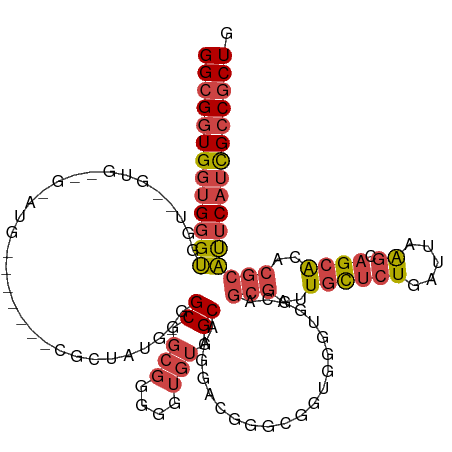
| Location | 20,283,677 – 20,283,790 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 123 |
| Reading direction | reverse |
| Mean pairwise identity | 68.86 |
| Shannon entropy | 0.57685 |
| G+C content | 0.64404 |
| Mean single sequence MFE | -47.00 |
| Consensus MFE | -24.08 |
| Energy contribution | -25.45 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.988781 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 20283677 113 - 27905053 GGCGGUGGUGGGUGGUAAGUGCGGCAUG-GACCCACGCUAUG-GGCGGCGGGGGUGUGGCAAGGAC--------GGUGGCAGCGAUUGCUCUGAUUAAGCAGCACACGCAUUCAUCGCCGCUG ((((((((((((((((...(((.((((.-..(((.(((....-.)))..))).)))).)))...))--------.(((((.((((((.....))))..)).)).))).)))))))))))))). ( -50.30, z-score = -1.79, R) >droSim1.chr3R 20124792 103 - 27517382 GGCGGUGGUGGGUGGC-------------------CGCUAUG-GGCGGCGGGGGUGUGGCAAGGACGGGCGGUGGGUGGCAGCGAUUGCUCUGAUUAAGCAGCACACGCAUUCAUCGCCGCUG ((((((((((((((((-------------------(((....-.)))))....(((((((...((((((((((.(.......).))))).))).))..))..))))).)))))))))))))). ( -48.00, z-score = -2.21, R) >droSec1.super_0 20620270 103 - 21120651 GGCGGUGGUGGGUGGC-------------------CGCUAUG-GGCGGCGGGGGUGUGGCAAGGACGGGCGGUGGGUGGCAGCGAUUGCUCUGAUUAAGCAGCACACGCAUUCAUCGCCGCUG ((((((((((((((((-------------------(((....-.)))))....(((((((...((((((((((.(.......).))))).))).))..))..))))).)))))))))))))). ( -48.00, z-score = -2.21, R) >droYak2.chr3R 2612712 117 + 28832112 GGCGGUGGUGGGUGGUGAUUGCGGCAUG-G----GCGGUAUG-GGCGGCGGGGGUGUGGCAAGGACGGGCGGUGGGUGGCAGCGAUUGCUCUGAUUAAGGAGCACACGCAUUCAUCGCCGCUG (((((((((((((((((((((((.(((.-.----((.((.((-.((.(((....))).)).....)).)).))..))).).)))))((((((......))))))))).)))))))))))))). ( -51.50, z-score = -3.33, R) >droEre2.scaffold_4820 2694459 123 + 10470090 GGCGGUGGUGGGUGGUGGGUGUUGUGUGCGGCAUGGGCGGUACGGCAGCGGGGGUGUGGCAAGGGCGGGCGGUGGGUGGCAGCGAUUGCUCUGAUUAAGCAGCACACGCAUUCAGCGCCGCUG (((((((.(((((((((..((((((((((.((....)).)))).))))))..(.(((....(((((((.((.((.....)).)).)))))))......))).).))).)))))).))))))). ( -54.20, z-score = -2.07, R) >droVir3.scaffold_13047 3647464 106 - 19223366 GGGGUUGGAGGGCAAGAGGCGUCGGAUG------AGAAAGAACAGCAACAAGAGCAAAGCAAAGG--------AGAUUCCCAAAAUUUUUGUUUUUGGGCUGCG---GCGCACAUUGUCGCUG ........(((((((...((((((....------........((((..(((((((((((....((--------......)).....))))))))))).))))))---))))...))))).)). ( -30.00, z-score = -0.65, R) >consensus GGCGGUGGUGGGUGGU__GUG__G_AUG_______CGCUAUG_GGCGGCGGGGGUGUGGCAAGGACGGGCGGUGGGUGGCAGCGAUUGCUCUGAUUAAGCAGCACACGCAUUCAUCGCCGCUG (((((((((((((...............................((.(((....))).)).....................(((..((((((.....)).))))..)))))))))))))))). (-24.08 = -25.45 + 1.37)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:40:43 2011