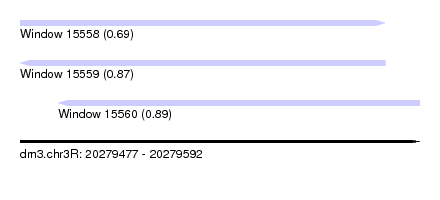
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,279,477 – 20,279,592 |
| Length | 115 |
| Max. P | 0.889088 |

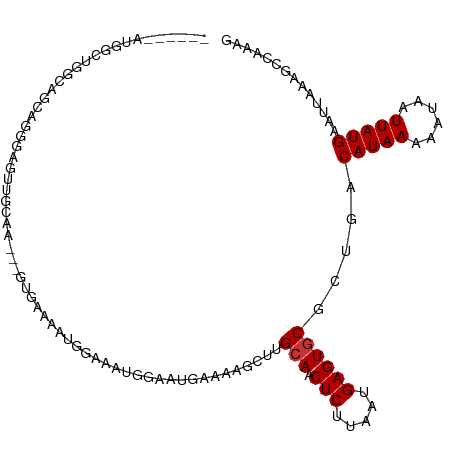
| Location | 20,279,477 – 20,279,582 |
|---|---|
| Length | 105 |
| Sequences | 8 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 77.59 |
| Shannon entropy | 0.44266 |
| G+C content | 0.36034 |
| Mean single sequence MFE | -23.61 |
| Consensus MFE | -7.46 |
| Energy contribution | -7.59 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.688769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 20279477 105 + 27905053 ------AUGGCUGGUAGCAGGGAGUUGCAA---GUGAAAAUGGAAAUGGAAUGAAAAGCUUGCAACUCUUAAUGAGUGCGCUGACAUAAAAAUAAUUAUGAAUUAAAGCCAAAG ------.((((((((.((((((((((((((---((....((.........)).....)))))))))))))......))))))..(((((......)))))......)))))... ( -29.30, z-score = -3.79, R) >droSim1.chr3R 20120535 105 + 27517382 ------AUGGCUGGCAGCAGGGAGUUGCAA---GUGAAAAUGGAAAUGGAAUGAAAAGCUUGCAACUCUUAAUGAGUGCGCUGACAUAAAAAUAAUUAUGAAUUAAAGCCAAAG ------.((((((((.((((((((((((((---((....((.........)).....)))))))))))))......))))))..(((((......)))))......)))))... ( -31.20, z-score = -3.97, R) >droSec1.super_0 20616008 105 + 21120651 ------AUGGCUGGCAGCAGGGAGUUGCAA---GUGAAAAUGGAAAUGGAAUGAAAAGCUAGCAACUCUUAAUGAGUGCGCUGACAUAAAAAUAAUUAUGAAUUAAAGCCAAAG ------.((((((((.((((((((((((.(---((....((.........)).....))).)))))))))......))))))..(((((......)))))......)))))... ( -27.10, z-score = -2.67, R) >droYak2.chr3R 2608302 105 - 28832112 ------GAAACUGGGAGUUGUGAGUUGGAA---GUGAAAAUGGAAAUGGAAUGAAAAGCUUGCAACUCUUAAUGAGUGCGCUGACAUAAAAAUAAUUAUGAAUUAAAGCCCAAG ------.....(((((((((..((((....---((....((....))...))....))))..))))))))).....((.(((..(((((......)))))......))).)).. ( -23.20, z-score = -2.96, R) >droEre2.scaffold_4820 2690277 105 - 10470090 ------GGAGCAGGCAGCUGGGAGUUGGAA---GUGAAAAUGGAAAUGGAAUGAAAAGCUUGCAACUCCUAAUGAGUGCGCUGACAUAAAAAUAAUUAUGAAUUAAAGCCAAAG ------((.(((..((..(((((((((.((---((....((.........)).....)))).))))))))).))..))).....(((((......)))))........)).... ( -27.60, z-score = -3.18, R) >droAna3.scaffold_13088 321345 100 + 569066 ---------AUAUACGGGAAUAAUAUACAA---A-AAAAAUCUAAAGCU-CAGAAACAAAUGCAACUCUUAAUGAGUGCGCUGACAUAAAAAUAAUUAUGAAUUAAAGCCAAAG ---------.......((..((((......---.-.............(-(((.......((((.(((.....)))))))))))(((((......))))).))))...)).... ( -12.01, z-score = -0.94, R) >droPer1.super_3 6121677 107 - 7375914 ------GUGAC-GAAAAUUGGGAAUGGGAAAAUGGGGGGAUGAAAAGCGCCAGCUACAAAUGCAACUCUUAAUGAGUGCGCUGACAUAAAAAUAAUUAUGAAUUAAAACCAAAG ------.....-.....((((........................(((((..((.......)).((((.....)))))))))..(((((......)))))........)))).. ( -19.00, z-score = -1.08, R) >droWil1.scaffold_181108 287150 111 - 4707319 GGACCAUGGUCCUGGUAAAACAAAAUGAAA---AUAAAAAUGCAAAGUAACGGCAACAACUGAAACUCUUAAUGAGUGCGCUGACAUAAAAAUAAUUAUGAAUUAAAACCAAAG ((((....))))((((..............---.................((((..(....)..((((.....))))..)))).(((((......))))).......))))... ( -19.50, z-score = -2.54, R) >consensus ______AUGGCUGGCAGCAGGGAGUUGCAA___GUGAAAAUGGAAAUGGAAUGAAAAGCUUGCAACUCUUAAUGAGUGCGCUGACAUAAAAAUAAUUAUGAAUUAAAGCCAAAG .............................................................(((.(((.....)))))).....(((((......))))).............. ( -7.46 = -7.59 + 0.12)

| Location | 20,279,477 – 20,279,582 |
|---|---|
| Length | 105 |
| Sequences | 8 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 77.59 |
| Shannon entropy | 0.44266 |
| G+C content | 0.36034 |
| Mean single sequence MFE | -17.94 |
| Consensus MFE | -9.00 |
| Energy contribution | -8.94 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.868933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 20279477 105 - 27905053 CUUUGGCUUUAAUUCAUAAUUAUUUUUAUGUCAGCGCACUCAUUAAGAGUUGCAAGCUUUUCAUUCCAUUUCCAUUUUCAC---UUGCAACUCCCUGCUACCAGCCAU------ ...(((((......(((((......)))))................((((((((((........................)---))))))))).........))))).------ ( -21.16, z-score = -3.56, R) >droSim1.chr3R 20120535 105 - 27517382 CUUUGGCUUUAAUUCAUAAUUAUUUUUAUGUCAGCGCACUCAUUAAGAGUUGCAAGCUUUUCAUUCCAUUUCCAUUUUCAC---UUGCAACUCCCUGCUGCCAGCCAU------ ...(((((......(((((......))))).(((((..........((((((((((........................)---)))))))))..)))))..))))).------ ( -23.26, z-score = -3.49, R) >droSec1.super_0 20616008 105 - 21120651 CUUUGGCUUUAAUUCAUAAUUAUUUUUAUGUCAGCGCACUCAUUAAGAGUUGCUAGCUUUUCAUUCCAUUUCCAUUUUCAC---UUGCAACUCCCUGCUGCCAGCCAU------ ...(((((......(((((......))))).(((((..........(((((((.((........................)---).)))))))..)))))..))))).------ ( -19.16, z-score = -1.94, R) >droYak2.chr3R 2608302 105 + 28832112 CUUGGGCUUUAAUUCAUAAUUAUUUUUAUGUCAGCGCACUCAUUAAGAGUUGCAAGCUUUUCAUUCCAUUUCCAUUUUCAC---UUCCAACUCACAACUCCCAGUUUC------ .(((((........(((((......)))))................((((((.(((........................)---)).))))))......)))))....------ ( -14.36, z-score = -2.18, R) >droEre2.scaffold_4820 2690277 105 + 10470090 CUUUGGCUUUAAUUCAUAAUUAUUUUUAUGUCAGCGCACUCAUUAGGAGUUGCAAGCUUUUCAUUCCAUUUCCAUUUUCAC---UUCCAACUCCCAGCUGCCUGCUCC------ .....(((......(((((......)))))..)))(((..((((.(((((((.(((........................)---)).))))))).)).))..)))...------ ( -17.16, z-score = -1.50, R) >droAna3.scaffold_13088 321345 100 - 569066 CUUUGGCUUUAAUUCAUAAUUAUUUUUAUGUCAGCGCACUCAUUAAGAGUUGCAUUUGUUUCUG-AGCUUUAGAUUUUU-U---UUGUAUAUUAUUCCCGUAUAU--------- ((..(((((.....(((((......)))))...(((.((((.....)))))))..........)-))))..))......-.---..((((((.......))))))--------- ( -15.20, z-score = -0.84, R) >droPer1.super_3 6121677 107 + 7375914 CUUUGGUUUUAAUUCAUAAUUAUUUUUAUGUCAGCGCACUCAUUAAGAGUUGCAUUUGUAGCUGGCGCUUUUCAUCCCCCCAUUUUCCCAUUCCCAAUUUUC-GUCAC------ ..((((........(((((......)))))..(((((.((.....))((((((....)))))).)))))........................)))).....-.....------ ( -16.80, z-score = -2.12, R) >droWil1.scaffold_181108 287150 111 + 4707319 CUUUGGUUUUAAUUCAUAAUUAUUUUUAUGUCAGCGCACUCAUUAAGAGUUUCAGUUGUUGCCGUUACUUUGCAUUUUUAU---UUUCAUUUUGUUUUACCAGGACCAUGGUCC ...((((...(((.(((((......))))).((((..((((.....))))....)))).......................---.........)))..))))((((....)))) ( -16.40, z-score = -0.69, R) >consensus CUUUGGCUUUAAUUCAUAAUUAUUUUUAUGUCAGCGCACUCAUUAAGAGUUGCAAGCUUUUCAUUCCAUUUCCAUUUUCAC___UUGCAACUCCCUGCUGCCAGCCAC______ .....(((......(((((......)))))..)))((((((.....))).)))............................................................. ( -9.00 = -8.94 + -0.06)

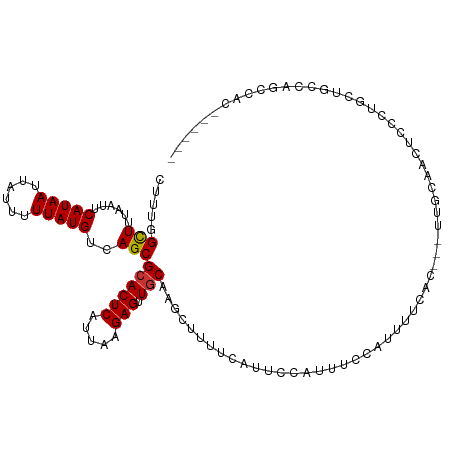

| Location | 20,279,488 – 20,279,592 |
|---|---|
| Length | 104 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 78.01 |
| Shannon entropy | 0.44468 |
| G+C content | 0.33204 |
| Mean single sequence MFE | -15.85 |
| Consensus MFE | -9.04 |
| Energy contribution | -8.78 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.889088 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 20279488 104 - 27905053 AACC-AAACAACUUUGGCUUUAAUUCAUAAUUAUUUUUAUGUCAGCGCACUCAUUAAGAGUUGCAAGCUUUUCAUUCCAUUUCCAUUUUCAC---UUGCAACUCCCUG------ ..((-(((....)))))........(((((......)))))................((((((((((........................)---)))))))))....------ ( -17.36, z-score = -3.04, R) >droSim1.chr3R 20120546 104 - 27517382 AACC-AAACAACUUUGGCUUUAAUUCAUAAUUAUUUUUAUGUCAGCGCACUCAUUAAGAGUUGCAAGCUUUUCAUUCCAUUUCCAUUUUCAC---UUGCAACUCCCUG------ ..((-(((....)))))........(((((......)))))................((((((((((........................)---)))))))))....------ ( -17.36, z-score = -3.04, R) >droSec1.super_0 20616019 104 - 21120651 AACC-AAACAACUUUGGCUUUAAUUCAUAAUUAUUUUUAUGUCAGCGCACUCAUUAAGAGUUGCUAGCUUUUCAUUCCAUUUCCAUUUUCAC---UUGCAACUCCCUG------ ..((-(((....)))))........(((((......)))))................(((((((.((........................)---).)))))))....------ ( -13.26, z-score = -1.41, R) >droYak2.chr3R 2608313 105 + 28832112 AACCAAAACAACUUGGGCUUUAAUUCAUAAUUAUUUUUAUGUCAGCGCACUCAUUAAGAGUUGCAAGCUUUUCAUUCCAUUUCCAUUUUCAC---UUCCAACUCACAA------ ..((((......))))((((.....(((((......)))))...(((.((((.....)))))))))))........................---.............------ ( -14.70, z-score = -2.46, R) >droEre2.scaffold_4820 2690288 104 + 10470090 AACC-AAACAACUUUGGCUUUAAUUCAUAAUUAUUUUUAUGUCAGCGCACUCAUUAGGAGUUGCAAGCUUUUCAUUCCAUUUCCAUUUUCAC---UUCCAACUCCCAG------ ..((-(((....)))))........(((((......)))))...............(((((((.(((........................)---)).)))))))...------ ( -14.06, z-score = -2.26, R) >droAna3.scaffold_13088 321356 99 - 569066 AACC-AGGCAACUUUGGCUUUAAUUCAUAAUUAUUUUUAUGUCAGCGCACUCAUUAAGAGUUGCAUUUGUUUCUGAGCUUUAGAUUUUUUUU---GUAUAUUA----------- ....-......((..(((((.....(((((......)))))...(((.((((.....)))))))..........)))))..)).........---........----------- ( -15.30, z-score = 0.03, R) >droPer1.super_3 6121687 107 + 7375914 AACU-AAACAACUUUGGUUUUAAUUCAUAAUUAUUUUUAUGUCAGCGCACUCAUUAAGAGUUGCAUUUGUAGCUGGCGCUUUUCAUCCCCCCAUUUUCCCAUUCCCAA------ ....-........((((........(((((......)))))..(((((.((.....))((((((....)))))).)))))........................))))------ ( -16.20, z-score = -1.46, R) >droWil1.scaffold_181108 287161 110 + 4707319 AACA-AACCAACUUUGGUUUUAAUUCAUAAUUAUUUUUAUGUCAGCGCACUCAUUAAGAGUUUCAGUUGUUGCCGUUACUUUGCAUUUUUAU---UUUCAUUUUGUUUUACCAG ((((-((........(((.......(((((......))))).((((..((((.....))))....))))..)))((......))........---......))))))....... ( -12.50, z-score = -0.30, R) >droGri2.scaffold_14906 297385 96 - 14172833 AACU-GAACAACUUGGGCUUUAAUUCAUAAUUAUUUUUAUGUCAGCGCAUUCAUU-AGAAUCGCUGUUGUUGUUCUUGCUGU---UGUUUAU---AUGCAUUUA---------- ...(-(((((((..((((..(((..(((((......))))).(((((.((((...-.))))))))))))..)))).....))---)))))).---.........---------- ( -21.90, z-score = -2.29, R) >consensus AACC_AAACAACUUUGGCUUUAAUUCAUAAUUAUUUUUAUGUCAGCGCACUCAUUAAGAGUUGCAAGCUUUUCAUUCCAUUUCCAUUUUCAC___UUGCAACUCCCUG______ ................(((......(((((......)))))..)))((((((.....)))).)).................................................. ( -9.04 = -8.78 + -0.26)

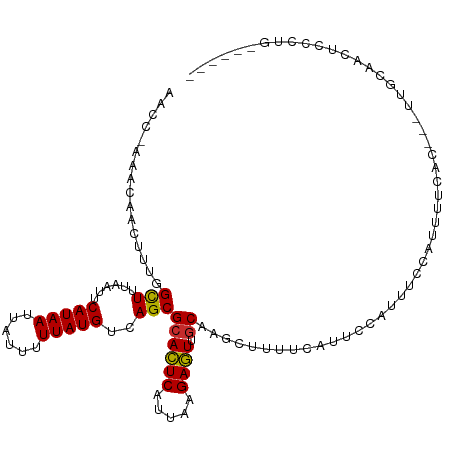
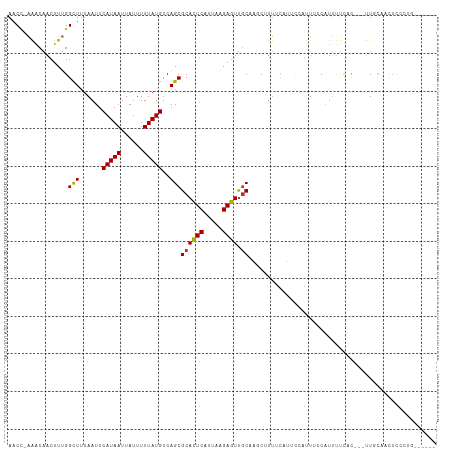
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:40:40 2011