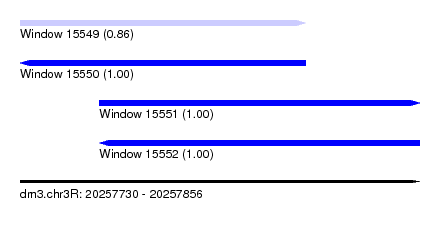
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,257,730 – 20,257,856 |
| Length | 126 |
| Max. P | 0.998668 |

| Location | 20,257,730 – 20,257,820 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 77.76 |
| Shannon entropy | 0.39338 |
| G+C content | 0.33294 |
| Mean single sequence MFE | -18.83 |
| Consensus MFE | -9.15 |
| Energy contribution | -9.99 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.858195 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
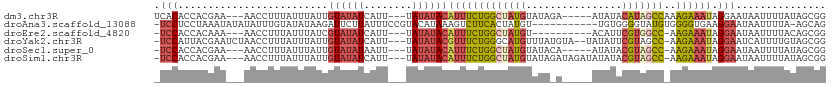
>dm3.chr3R 20257730 90 + 27905053 CAUUUGAAACUUGGACAACAGAUGUCCGCUAUAAAAUUAUUCCUAUUUCUUUGGCUAUGUAUAU-----UCUAUACAUAGCCAGAAAUGUAUAUA--------- ............(((((.....)))))................(((((((..(((((((((((.-----..))))))))))))))))))......--------- ( -24.70, z-score = -4.55, R) >droAna3.scaffold_13088 297639 92 + 569066 CAUUUGAAACUUGGACAACAGAUGUCUGCU-UAAAAUUAUUCCUUCACCCCACAUACCCCACA-----------ACAUAGUGAAGACUUUAUGUACGGAAAUAA ..((((......(((((.....)))))...-...........((((((...............-----------.....))))))..........))))..... ( -9.55, z-score = 0.82, R) >droEre2.scaffold_4820 2667960 84 - 10470090 CAUUUGAAAGUUGGACAACAGAUGUCCGCUGUAAAAUUAUUCCUAUUUCUU-GGCCACGAAUGU----------ACAUAGCCAGAAAUGUAUAUA--------- .((((...(((.(((((.....))))))))...))))......(((((((.-(((.........----------.....))))))))))......--------- ( -16.24, z-score = -1.03, R) >droYak2.chr3R 2585911 92 - 28832112 CAUUUGAAAGUUGGACAACAGAUGUCCGCUACAAAAUGAUUCCUAUUUCUU-GGCUACGAAUAUAU--ACAUAAACAUGCCCAGAAACGUAUAUA--------- (((((...(((.(((((.....))))))))...))))).......(((((.-(((...........--..........))).)))))........--------- ( -17.60, z-score = -1.94, R) >droSec1.super_0 20594718 89 + 21120651 CAUUUGAAACUUGGACAACAGAUGUCCGCUAUAAAAUUAUUCCUAUUUCUU-GGCUACGUAUAU-----UGUAUACAUAGCCAGAAAUGUAUAUA--------- ............(((((.....)))))................(((((((.-(((((.(((((.-----..))))).))))))))))))......--------- ( -22.00, z-score = -3.39, R) >droSim1.chr3R 20095877 94 + 27517382 CAUUUGAAACUUGGACAACAGAUGUCCGCUAUAAAAUUAUUCCUAUUUCUU-GGCUACGUAUAUAUCUAUCUAUACAUAGCCAGAAAUGUAUAUA--------- ............(((((.....)))))................(((((((.-(((((.(((((........))))).))))))))))))......--------- ( -22.90, z-score = -3.78, R) >consensus CAUUUGAAACUUGGACAACAGAUGUCCGCUAUAAAAUUAUUCCUAUUUCUU_GGCUACGUAUAU_____U_UAUACAUAGCCAGAAAUGUAUAUA_________ ............(((((.....)))))................(((((((..(((((.(................).))))))))))))............... ( -9.15 = -9.99 + 0.84)
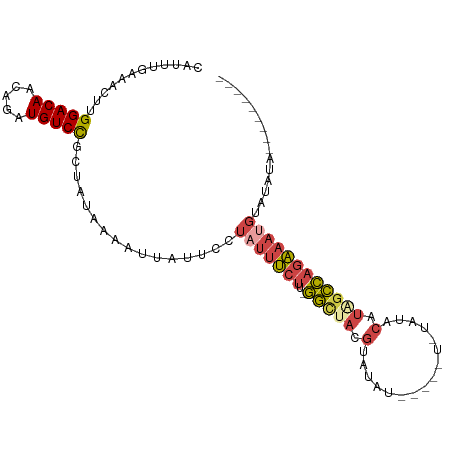
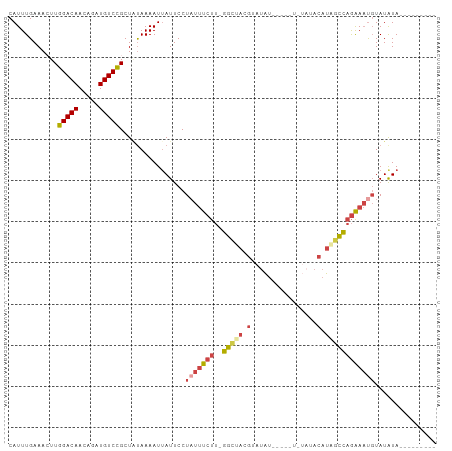
| Location | 20,257,730 – 20,257,820 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 77.76 |
| Shannon entropy | 0.39338 |
| G+C content | 0.33294 |
| Mean single sequence MFE | -23.97 |
| Consensus MFE | -14.58 |
| Energy contribution | -14.76 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.44 |
| Mean z-score | -3.35 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.35 |
| SVM RNA-class probability | 0.998414 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 20257730 90 - 27905053 ---------UAUAUACAUUUCUGGCUAUGUAUAGA-----AUAUACAUAGCCAAAGAAAUAGGAAUAAUUUUAUAGCGGACAUCUGUUGUCCAAGUUUCAAAUG ---------......(((((.((((((((((((..-----.))))))))))))..(((((..(((....))).....(((((.....)))))..)))))))))) ( -25.50, z-score = -4.45, R) >droAna3.scaffold_13088 297639 92 - 569066 UUAUUUCCGUACAUAAAGUCUUCACUAUGU-----------UGUGGGGUAUGUGGGGUGAAGGAAUAAUUUUA-AGCAGACAUCUGUUGUCCAAGUUUCAAAUG ...((.((.((((((....((((((.....-----------.)))))))))))).)).)).(((((.......-(((((....)))))......)))))..... ( -20.92, z-score = -1.17, R) >droEre2.scaffold_4820 2667960 84 + 10470090 ---------UAUAUACAUUUCUGGCUAUGU----------ACAUUCGUGGCC-AAGAAAUAGGAAUAAUUUUACAGCGGACAUCUGUUGUCCAACUUUCAAAUG ---------.......(((((((((((((.----------.....)))))))-.))))))..(((.......(((((((....)))))))......)))..... ( -23.32, z-score = -3.79, R) >droYak2.chr3R 2585911 92 + 28832112 ---------UAUAUACGUUUCUGGGCAUGUUUAUGU--AUAUAUUCGUAGCC-AAGAAAUAGGAAUCAUUUUGUAGCGGACAUCUGUUGUCCAACUUUCAAAUG ---------.......((((((.((((((..(((..--..)))..))).)))-.))))))......(((((...((.(((((.....)))))..))...))))) ( -19.60, z-score = -1.31, R) >droSec1.super_0 20594718 89 - 21120651 ---------UAUAUACAUUUCUGGCUAUGUAUACA-----AUAUACGUAGCC-AAGAAAUAGGAAUAAUUUUAUAGCGGACAUCUGUUGUCCAAGUUUCAAAUG ---------.......(((((((((((((((((..-----.)))))))))))-.)))))).(((((......(((((((....)))))))....)))))..... ( -26.40, z-score = -4.77, R) >droSim1.chr3R 20095877 94 - 27517382 ---------UAUAUACAUUUCUGGCUAUGUAUAGAUAGAUAUAUACGUAGCC-AAGAAAUAGGAAUAAUUUUAUAGCGGACAUCUGUUGUCCAAGUUUCAAAUG ---------.......(((((((((((((((((........)))))))))))-.)))))).(((((......(((((((....)))))))....)))))..... ( -28.10, z-score = -4.61, R) >consensus _________UAUAUACAUUUCUGGCUAUGUAUA_A_____AUAUACGUAGCC_AAGAAAUAGGAAUAAUUUUAUAGCGGACAUCUGUUGUCCAAGUUUCAAAUG ................(((((((((((((................)))))))..)))))).(((((......(((((((....)))))))....)))))..... (-14.58 = -14.76 + 0.17)
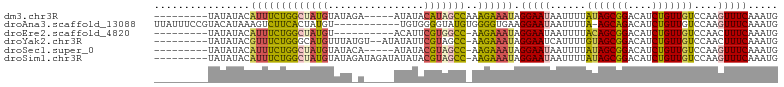
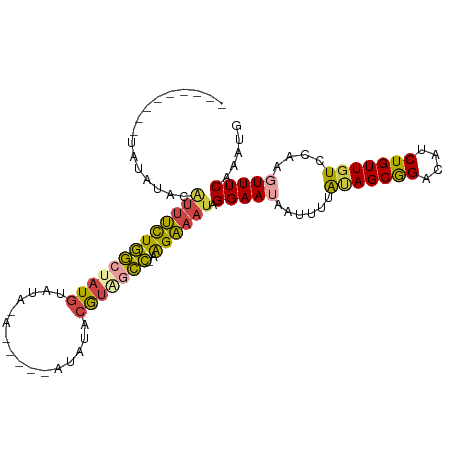
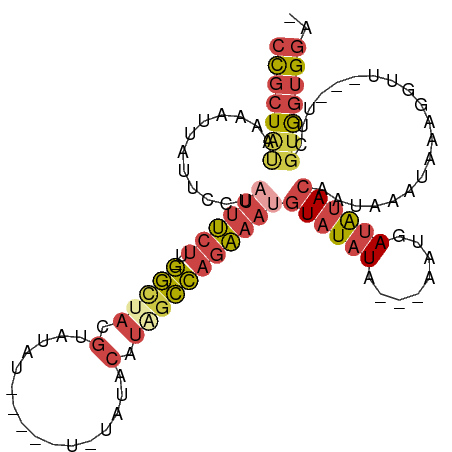
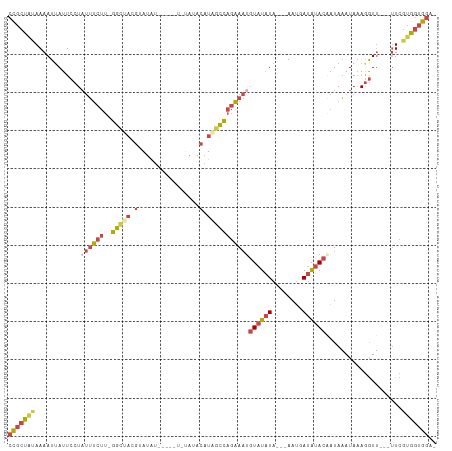
| Location | 20,257,755 – 20,257,856 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 72.66 |
| Shannon entropy | 0.49943 |
| G+C content | 0.30105 |
| Mean single sequence MFE | -24.06 |
| Consensus MFE | -10.03 |
| Energy contribution | -11.87 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.36 |
| Mean z-score | -3.18 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.85 |
| SVM RNA-class probability | 0.995797 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 20257755 101 + 27905053 CCGCUAUAAAAUUAUUCCUAUUUCUUUGGCUAUGUAUAU-----UCUAUACAUAGCCAGAAAUGUAUAUA---AAUGAUAUACAAUAAAUAAAGGUU---UUCGUGGUGUGA .(((((..(((.....(((.....((((((((((((((.-----..))))))))))))))..(((((((.---....)))))))........))).)---))..)))))... ( -29.90, z-score = -5.13, R) >droAna3.scaffold_13088 297664 99 + 569066 CUGCU-UAAAAUUAUUCCUUCACCCCACAUACCCCACA-----------ACAUAGUGAAGACUUUAUGUACGGAAAUAAGAAUCUUAUAUACAAAUAUAUAUUUAGGAGGA- .....-.........((((((.((..((((((..(((.-----------.....)))..)....)))))..))............((((((....))))))....))))))- ( -13.20, z-score = -0.38, R) >droEre2.scaffold_4820 2667985 94 - 10470090 CCGCUGUAAAAUUAUUCCUAUUUCUU-GGCCACGAAUGU----------ACAUAGCCAGAAAUGUAUAUA---AAUGAUAUACGAUAAAUAAAGGUU---UUUGUGGUGGA- ((((..(((((.....(((((((((.-(((.........----------.....)))))))))((((((.---....)))))).........))).)---))))..)))).- ( -23.74, z-score = -2.69, R) >droYak2.chr3R 2585936 105 - 28832112 CCGCUACAAAAUGAUUCCUAUUUCUU-GGCUACGAAUAUA--UACAUAAACAUGCCCAGAAACGUAUAUA---AAUGAUAUACAAUAAAUAAAGGUUAGAUUCGUAAUGGA- (((.(((.........(((.(((((.-(((..........--...........))).))))).((((((.---....)))))).........)))........))).))).- ( -16.93, z-score = -1.02, R) >droSec1.super_0 20594743 99 + 21120651 CCGCUAUAAAAUUAUUCCUAUUUCUU-GGCUACGUAUAU-----UGUAUACAUAGCCAGAAAUGUAUAUA---AAUUAUAUACAAUAAAUAAAGGUU---UUCGUGGUGGA- ((((((..(((.....(((((((((.-(((((.(((((.-----..))))).)))))))))))(((((((---...))))))).........))).)---))..)))))).- ( -29.90, z-score = -4.88, R) >droSim1.chr3R 20095902 104 + 27517382 CCGCUAUAAAAUUAUUCCUAUUUCUU-GGCUACGUAUAUAUCUAUCUAUACAUAGCCAGAAAUGUAUAUA---AAUGAUAUACAAUAAAUAAAGGUU---UUCGUGGUGGA- ((((((..(((.....(((((((((.-(((((.(((((........))))).)))))))))))((((((.---....)))))).........))).)---))..)))))).- ( -30.70, z-score = -5.00, R) >consensus CCGCUAUAAAAUUAUUCCUAUUUCUU_GGCUACGUAUAU_____U_UAUACAUAGCCAGAAAUGUAUAUA___AAUGAUAUACAAUAAAUAAAGGUU___UUCGUGGUGGA_ (((((((.........(((((((((..(((((.(................).)))))))))))((((((........)))))).........)))........))))))).. (-10.03 = -11.87 + 1.84)
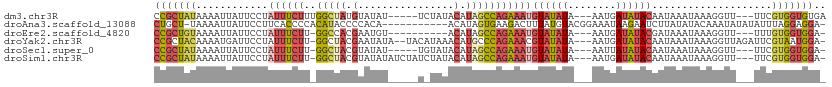
| Location | 20,257,755 – 20,257,856 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 72.66 |
| Shannon entropy | 0.49943 |
| G+C content | 0.30105 |
| Mean single sequence MFE | -23.45 |
| Consensus MFE | -10.96 |
| Energy contribution | -12.02 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.36 |
| Mean z-score | -3.34 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.44 |
| SVM RNA-class probability | 0.998668 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
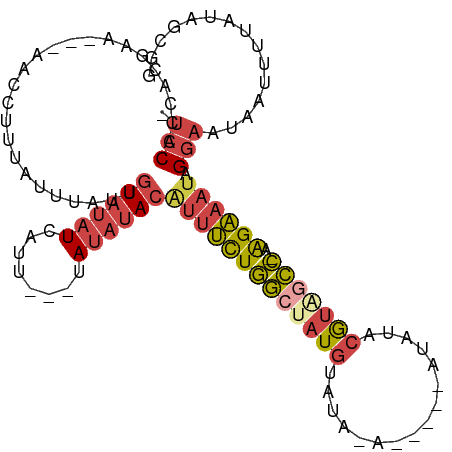
>dm3.chr3R 20257755 101 - 27905053 UCACACCACGAA---AACCUUUAUUUAUUGUAUAUCAUU---UAUAUACAUUUCUGGCUAUGUAUAGA-----AUAUACAUAGCCAAAGAAAUAGGAAUAAUUUUAUAGCGG ........((..---.....((((((...((((((....---.))))))(((((((((((((((((..-----.)))))))))))..))))))..))))))........)). ( -24.06, z-score = -4.55, R) >droAna3.scaffold_13088 297664 99 - 569066 -UCCUCCUAAAUAUAUAUUUGUAUAUAAGAUUCUUAUUUCCGUACAUAAAGUCUUCACUAUGU-----------UGUGGGGUAUGUGGGGUGAAGGAAUAAUUUUA-AGCAG -..........((((((....))))))..((((((.((.((.((((((....((((((.....-----------.)))))))))))).)).)))))))).......-..... ( -19.60, z-score = -0.46, R) >droEre2.scaffold_4820 2667985 94 + 10470090 -UCCACCACAAA---AACCUUUAUUUAUCGUAUAUCAUU---UAUAUACAUUUCUGGCUAUGU----------ACAUUCGUGGCC-AAGAAAUAGGAAUAAUUUUACAGCGG -(((........---..............((((((....---.))))))(((((((((((((.----------.....)))))))-.)))))).)))............... ( -21.60, z-score = -3.78, R) >droYak2.chr3R 2585936 105 + 28832112 -UCCAUUACGAAUCUAACCUUUAUUUAUUGUAUAUCAUU---UAUAUACGUUUCUGGGCAUGUUUAUGUA--UAUAUUCGUAGCC-AAGAAAUAGGAAUCAUUUUGUAGCGG -.((.(((((((.....(((.........((((((....---.))))))((((((.((((((..(((...--.)))..))).)))-.)))))))))......))))))).)) ( -22.50, z-score = -2.08, R) >droSec1.super_0 20594743 99 - 21120651 -UCCACCACGAA---AACCUUUAUUUAUUGUAUAUAAUU---UAUAUACAUUUCUGGCUAUGUAUACA-----AUAUACGUAGCC-AAGAAAUAGGAAUAAUUUUAUAGCGG -.......((..---.....((((((...(((((((...---)))))))(((((((((((((((((..-----.)))))))))))-.))))))..))))))........)). ( -25.96, z-score = -4.75, R) >droSim1.chr3R 20095902 104 - 27517382 -UCCACCACGAA---AACCUUUAUUUAUUGUAUAUCAUU---UAUAUACAUUUCUGGCUAUGUAUAGAUAGAUAUAUACGUAGCC-AAGAAAUAGGAAUAAUUUUAUAGCGG -.......((..---.....((((((...((((((....---.))))))(((((((((((((((((........)))))))))))-.))))))..))))))........)). ( -26.96, z-score = -4.42, R) >consensus _UCCACCACGAA___AACCUUUAUUUAUUGUAUAUCAUU___UAUAUACAUUUCUGGCUAUGUAUA_A_____AUAUACGUAGCC_AAGAAAUAGGAAUAAUUUUAUAGCGG .(((.........................((((((........))))))(((((((((((((................)))))))..)))))).)))............... (-10.96 = -12.02 + 1.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:40:33 2011