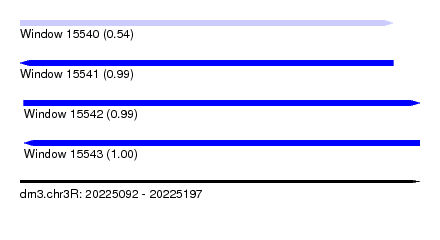
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,225,092 – 20,225,197 |
| Length | 105 |
| Max. P | 0.995889 |

| Location | 20,225,092 – 20,225,190 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 80.60 |
| Shannon entropy | 0.31748 |
| G+C content | 0.57780 |
| Mean single sequence MFE | -30.60 |
| Consensus MFE | -20.98 |
| Energy contribution | -21.23 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.543374 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
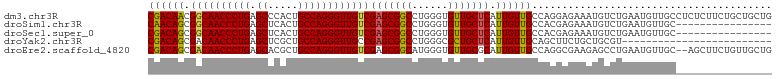
>dm3.chr3R 20225092 98 + 27905053 GCAGCAGCAGAAGAGAGGCAACAUUCAGACAUUUCUCCUGGCAACAAUGAGCAACACCCAGGCCGCUCGACAACCCUGGCAGUGGGCUCAGGGUUGCC ((....))....(((.(....).))).............((((((..(((((..((((((((............)))))..))).)))))..)))))) ( -30.80, z-score = -0.26, R) >droSim1.chr3R 20063208 81 + 27517382 -----------------GCAACAUUCAGACAUUUCUCGUGGCAACAAUGAGCAACACCCAGGCCGCUCGACAACCCUGGCAGUGAGCUCAGGGUUGCC -----------------......................((((((..(((((..((((((((............)))))..))).)))))..)))))) ( -25.90, z-score = -1.37, R) >droSec1.super_0 20563658 81 + 21120651 -----------------GCAACAUUCAGACAUUUCUCGUGGCAACAAUGAGCAACACCCAGGCCGCUCGACAACCCUGGCAGUGAGCUCAGGGUUGCC -----------------......................((((((..(((((..((((((((............)))))..))).)))))..)))))) ( -25.90, z-score = -1.37, R) >droEre2.scaffold_4820 2635281 96 - 10470090 --GCAGCAACAGAAGCUGCAACAUUCAGGCUCUUCGCCUGGCAACAAUGCGCAACACCCAUGCCGCUCGACAACCCUGGCAGCGUGCUCAGGGUUGUC --(((((.......))))).....((((((.....)))))).......(((((.......)).)))..((((((((((((.....)).)))))))))) ( -39.80, z-score = -3.01, R) >consensus _________________GCAACAUUCAGACAUUUCUCCUGGCAACAAUGAGCAACACCCAGGCCGCUCGACAACCCUGGCAGUGAGCUCAGGGUUGCC ......................................((....)).(((((............))))).((((((((((.....)).)))))))).. (-20.98 = -21.23 + 0.25)

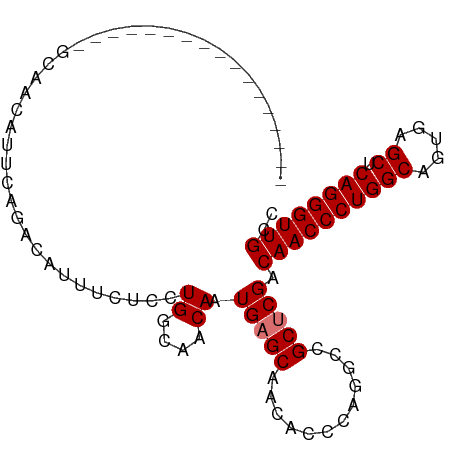
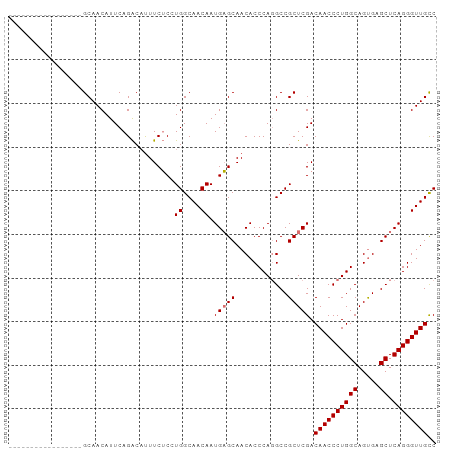
| Location | 20,225,092 – 20,225,190 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 80.60 |
| Shannon entropy | 0.31748 |
| G+C content | 0.57780 |
| Mean single sequence MFE | -37.65 |
| Consensus MFE | -31.81 |
| Energy contribution | -30.88 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.988354 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 20225092 98 - 27905053 GGCAACCCUGAGCCCACUGCCAGGGUUGUCGAGCGGCCUGGGUGUUGCUCAUUGUUGCCAGGAGAAAUGUCUGAAUGUUGCCUCUCUUCUGCUGCUGC ((((((((((.((.....))))))))))))(((((((......)))))))...((.((.((((((...(.(........).)..)))))))).))... ( -36.90, z-score = -1.19, R) >droSim1.chr3R 20063208 81 - 27517382 GGCAACCCUGAGCUCACUGCCAGGGUUGUCGAGCGGCCUGGGUGUUGCUCAUUGUUGCCACGAGAAAUGUCUGAAUGUUGC----------------- ((((((((((.((.....))))))))))))(((((((......)))))))......((.((((((....)))...))).))----------------- ( -31.60, z-score = -1.87, R) >droSec1.super_0 20563658 81 - 21120651 GGCAACCCUGAGCUCACUGCCAGGGUUGUCGAGCGGCCUGGGUGUUGCUCAUUGUUGCCACGAGAAAUGUCUGAAUGUUGC----------------- ((((((((((.((.....))))))))))))(((((((......)))))))......((.((((((....)))...))).))----------------- ( -31.60, z-score = -1.87, R) >droEre2.scaffold_4820 2635281 96 + 10470090 GACAACCCUGAGCACGCUGCCAGGGUUGUCGAGCGGCAUGGGUGUUGCGCAUUGUUGCCAGGCGAAGAGCCUGAAUGUUGCAGCUUCUGUUGCUGC-- ((((((((((.((.....))))))))))))..((((((((((.((((((((....)))(((((.....)))))......))))).)))).))))))-- ( -50.50, z-score = -4.35, R) >consensus GGCAACCCUGAGCUCACUGCCAGGGUUGUCGAGCGGCCUGGGUGUUGCUCAUUGUUGCCACGAGAAAUGUCUGAAUGUUGC_________________ ((((((((((.((.....))))))))))))..(((((.((((..((.(((...........))).))..))))...)))))................. (-31.81 = -30.88 + -0.94)

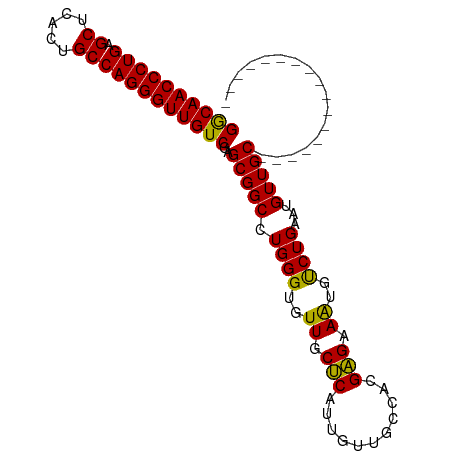
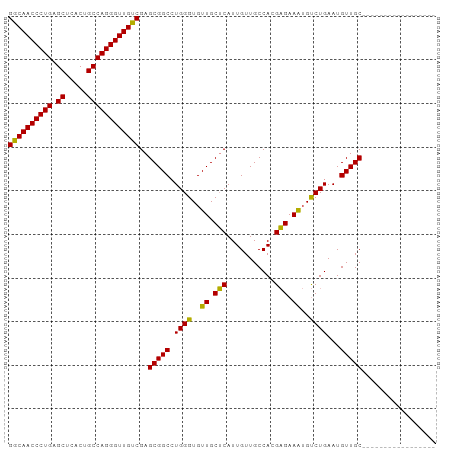
| Location | 20,225,093 – 20,225,197 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 76.06 |
| Shannon entropy | 0.39962 |
| G+C content | 0.59618 |
| Mean single sequence MFE | -37.38 |
| Consensus MFE | -25.68 |
| Energy contribution | -26.28 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.57 |
| SVM RNA-class probability | 0.992912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 20225093 104 + 27905053 CAGCAGCAGAAGAGAGGCAACAUUCAGACAUUUCUCCUGGCAACAAUGAGCAACACCCAGGCCGCUCGACAACCCUGGCAGUGGGCUCAGGGUUGCCGUUGUCG ..(((((....(((.(....).))).............((((((..(((((..((((((((............)))))..))).)))))..))))))))))).. ( -36.50, z-score = -1.34, R) >droSim1.chr3R 20063208 88 + 27517382 ----------------GCAACAUUCAGACAUUUCUCGUGGCAACAAUGAGCAACACCCAGGCCGCUCGACAACCCUGGCAGUGAGCUCAGGGUUGCCGCUGUUG ----------------.(((((....((......))((((((((..(((((..((((((((............)))))..))).)))))..))))))))))))) ( -33.50, z-score = -2.52, R) >droSec1.super_0 20563658 88 + 21120651 ----------------GCAACAUUCAGACAUUUCUCGUGGCAACAAUGAGCAACACCCAGGCCGCUCGACAACCCUGGCAGUGAGCUCAGGGUUGCCGCUGUCG ----------------..........((((......((((((((..(((((..((((((((............)))))..))).)))))..)))))))))))). ( -33.50, z-score = -2.64, R) >droYak2.chr3R 2552661 79 - 28832112 -------------------------ACGCAGCAGAAGCUGCAACAAUGAGCAGCGCCCAGGCCGCUCGGCAACCCUGGCAGCGAGCUCAGGGUUGUCGCUGUCG -------------------------..(((((....)))))......((.((((((....)).....((((((((((((.....)).)))))))))))))))). ( -39.00, z-score = -2.49, R) >droEre2.scaffold_4820 2635282 102 - 10470090 CAGCAACAGAAGCU--GCAACAUUCAGGCUCUUCGCCUGGCAACAAUGCGCAACACCCAUGCCGCUCGACAACCCUGGCAGCGUGCUCAGGGUUGUCGCUGUCG ((((....((.((.--(((.....(((((.....)))))(((....)))..........))).))))((((((((((((.....)).))))))))))))))... ( -44.40, z-score = -3.62, R) >consensus ________________GCAACAUUCAGACAUUUCUCGUGGCAACAAUGAGCAACACCCAGGCCGCUCGACAACCCUGGCAGUGAGCUCAGGGUUGCCGCUGUCG ..........................((((......((((((((..(((((..((((((((............)))))..))).)))))..)))))))))))). (-25.68 = -26.28 + 0.60)
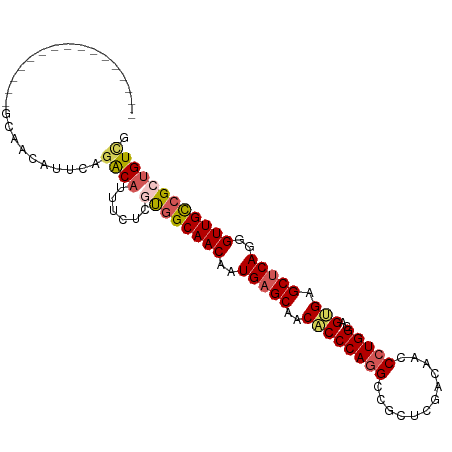
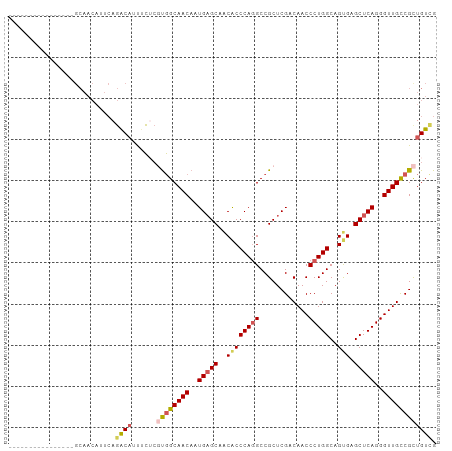
| Location | 20,225,093 – 20,225,197 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 76.06 |
| Shannon entropy | 0.39962 |
| G+C content | 0.59618 |
| Mean single sequence MFE | -40.68 |
| Consensus MFE | -33.88 |
| Energy contribution | -33.68 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.86 |
| SVM RNA-class probability | 0.995889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
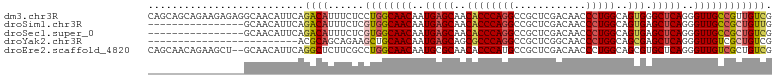
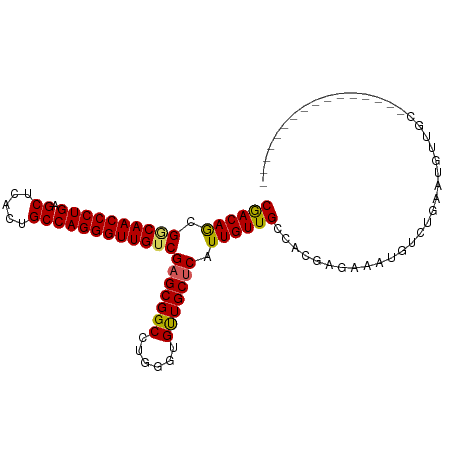
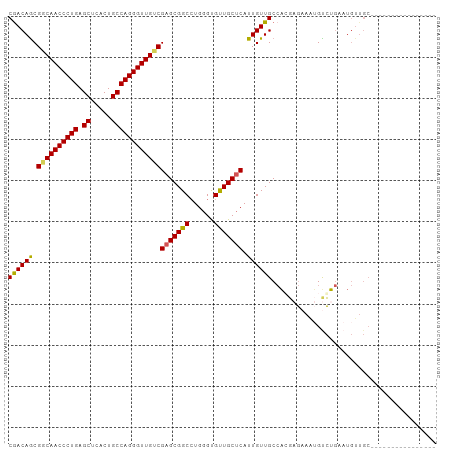
>dm3.chr3R 20225093 104 - 27905053 CGACAACGGCAACCCUGAGCCCACUGCCAGGGUUGUCGAGCGGCCUGGGUGUUGCUCAUUGUUGCCAGGAGAAAUGUCUGAAUGUUGCCUCUCUUCUGCUGCUG ((((((.((((((((((.((.....))))))))))))(((((((......))))))).)))))).(((((((...(.(........).)..)))))))...... ( -39.70, z-score = -1.78, R) >droSim1.chr3R 20063208 88 - 27517382 CAACAGCGGCAACCCUGAGCUCACUGCCAGGGUUGUCGAGCGGCCUGGGUGUUGCUCAUUGUUGCCACGAGAAAUGUCUGAAUGUUGC---------------- (((((((((((((((((.((.....))))))))))))(((((((......)))))))......))....(((....)))...))))).---------------- ( -36.00, z-score = -2.19, R) >droSec1.super_0 20563658 88 - 21120651 CGACAGCGGCAACCCUGAGCUCACUGCCAGGGUUGUCGAGCGGCCUGGGUGUUGCUCAUUGUUGCCACGAGAAAUGUCUGAAUGUUGC---------------- (((((((((((((((((.((.....))))))))))))(((((((......)))))))......))....(((....)))...))))).---------------- ( -35.70, z-score = -2.00, R) >droYak2.chr3R 2552661 79 + 28832112 CGACAGCGACAACCCUGAGCUCGCUGCCAGGGUUGCCGAGCGGCCUGGGCGCUGCUCAUUGUUGCAGCUUCUGCUGCGU------------------------- .(((((.(.((((((((.((.....)))))))))).)(((((((......))))))).)))))(((((....)))))..------------------------- ( -40.70, z-score = -2.52, R) >droEre2.scaffold_4820 2635282 102 + 10470090 CGACAGCGACAACCCUGAGCACGCUGCCAGGGUUGUCGAGCGGCAUGGGUGUUGCGCAUUGUUGCCAGGCGAAGAGCCUGAAUGUUGC--AGCUUCUGUUGCUG ......(((((((((((.((.....)))))))))))))(((((((.(((.((((((((....)))(((((.....)))))......))--))))))))))))). ( -51.30, z-score = -3.85, R) >consensus CGACAGCGGCAACCCUGAGCUCACUGCCAGGGUUGUCGAGCGGCCUGGGUGUUGCUCAUUGUUGCCACGAGAAAUGUCUGAAUGUUGC________________ ((((((.((((((((((.((.....))))))))))))(((((((......))))))).))))))........................................ (-33.88 = -33.68 + -0.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:40:26 2011