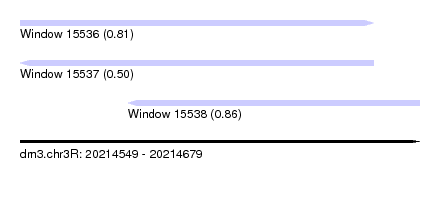
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,214,549 – 20,214,679 |
| Length | 130 |
| Max. P | 0.859308 |

| Location | 20,214,549 – 20,214,664 |
|---|---|
| Length | 115 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 80.44 |
| Shannon entropy | 0.43795 |
| G+C content | 0.31839 |
| Mean single sequence MFE | -13.38 |
| Consensus MFE | -10.14 |
| Energy contribution | -10.15 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.813322 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
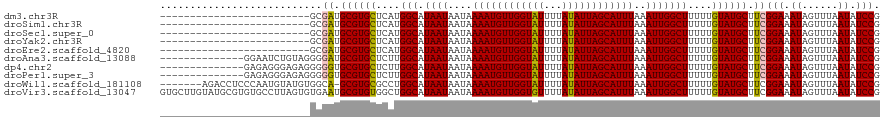
>dm3.chr3R 20214549 115 + 27905053 ACUUCAGACUCCCGACUCAGACGCACGGUCCUCUACGGAUAUUAAACUAUUUCCGAAGCAUACAAAAAGCCAAUUUAAAUGCUAAUAUAAAAUACCAACAUUUUAUUAUUAUGCC .(((((((...(((...........)))...)))..(((.((......)).)))))))......................(((((((((((((......)))))).))))).)). ( -13.30, z-score = -0.61, R) >droSim1.chr3R 20050708 115 + 27517382 ACUUCAGACUCGAGACUCAGACGCACGGUCCUCUACGGAUAUUAAACUAUUUCCGAAGCAUACAAAAAGCCAAUUUAAAUGCUAAUAUAAAAUACCAACAUUUUAUUAUUAUGCC .((((.(((.((.(.(......)).)))))......(((.((......)).)))))))......................(((((((((((((......)))))).))))).)). ( -13.50, z-score = -0.37, R) >droSec1.super_0 20553118 115 + 21120651 ACUUCAGACUUGAGACUCAGACGCACGGUCCUCUACGGAUAUUAAACUAUUUCCGAAGCAUACAAAAAGCCAAUUUAAAUGCUAAUAUAAAAUACCAACAUUUUAUUAUUAUGCC .((((......((((((.........))).)))...(((.((......)).)))))))......................(((((((((((((......)))))).))))).)). ( -13.10, z-score = -0.20, R) >droYak2.chr3R 2542145 113 - 28832112 ACCUC-GACUUC-GACUUCGACUCACGGUCCUCUACGGAUAUUAAACUAUUUCCGAAGCAUACAAAAAGCCAAUUUAAAUGCUAAUAUAAAAUACCAACAUUUUAUUAUUAUGCC .....-((((((-(....))).....)))).....((((.((......)).))))..(((((.....(((..........)))...(((((((......)))))))...))))). ( -14.50, z-score = -1.48, R) >droEre2.scaffold_4820 2624771 113 - 10470090 AGUGC-GACUCC-GACUCAGACUCACGGUCCUCUACGGAUAUUAAACUAUUUCCGAAGCAUACAAAAAGCCAAUUUAAAUGCUAAUAUAAAAUACCAACAUUUUAUUAUUAUGCC .((((-((((..-((.......))..)))).....((((.((......)).))))..))))...................(((((((((((((......)))))).))))).)). ( -15.90, z-score = -1.00, R) >droAna3.scaffold_13088 255142 110 + 569066 -ACUUAGACUCAGACUUGGACUAUUC----CUCUGCGGAUAUUAAACUAUUUCCGAAGCAUACAAAAAGCCAAUUUAAAUGCUAAUAUAAAAUACCAACAUUUUAUUAUUAUGCC -.........((((...(((....))----)))))((((.((......)).))))..(((((.....(((..........)))...(((((((......)))))))...))))). ( -13.80, z-score = -0.91, R) >dp4.chr2 12205452 115 - 30794189 AAGACAGACUCUGACUCUGAUAGUCCUCGCCCCUACGGAUAUUAAACUAUUUCCGAAGCAUACAAAAAGCCAAUUUAAAUGCUAAUAUAAAAUACCAACAUUUUAUUAUUAUGCC ....((((.......))))................((((.((......)).))))..(((((.....(((..........)))...(((((((......)))))))...))))). ( -12.90, z-score = -0.57, R) >droPer1.super_3 6055447 115 - 7375914 AAGACAGACUCUGACUCUGAUAGUCCUCGCCCCUACGGAUAUUAAACUAUUUCCGAAGCAUACAAAAAGCCAAUUUAAAUGCUAAUAUAAAAUACCAACAUUUUAUUAUUAUGCC ....((((.......))))................((((.((......)).))))..(((((.....(((..........)))...(((((((......)))))))...))))). ( -12.90, z-score = -0.57, R) >droWil1.scaffold_181108 216250 99 - 4707319 ----------------AAGAUCGGGGACUCUUUUUCGGAUAUUAAACUAUUUCCGAAGCAUACAAAAAGCCAAUUUAAAUGCUAAUAUAAAAUACCAACAUUUUAUUAUUAUGCC ----------------((((..(....))))).((((((.((......)).))))))(((((.....(((..........)))...(((((((......)))))))...))))). ( -14.10, z-score = -0.92, R) >droMoj3.scaffold_6540 20565350 89 + 34148556 --------------------------AAGACUUUUCGGAUAUUAAACUAUUUCCGAAGCAUACAAAAAGCCAAUUUAAAUGCUAAUAUAAAAUACCAACAUUUUAUUAUUAUGCC --------------------------.......((((((.((......)).))))))(((((.....(((..........)))...(((((((......)))))))...))))). ( -12.60, z-score = -2.33, R) >droVir3.scaffold_13047 3559562 89 + 19223366 --------------------------AAGACUUUUCGGAUAUUAAACUAUUUCCGAAGCAUACAAAAAGCCAAUUUAAAUGCUAAUAUAAAACACCAACAUUUUAUUAUUAUGCC --------------------------.......((((((.((......)).))))))(((((.....(((..........)))...((((((........))))))...))))). ( -11.10, z-score = -1.92, R) >droGri2.scaffold_14906 213847 89 + 14172833 --------------------------AAGACUUUUCGGAUAUUAAACUAUUUCCGAAGCAUACAAAAAGCCAAUUUAAAUGCUAAUAUAAAAUACCAACGUUUUAUUAUUAUGCC --------------------------.......((((((.((......)).))))))(((((.....(((..........)))...(((((((......)))))))...))))). ( -12.90, z-score = -1.98, R) >consensus A__UC_GACUC__GACUCAGACGCACGGUCCUCUACGGAUAUUAAACUAUUUCCGAAGCAUACAAAAAGCCAAUUUAAAUGCUAAUAUAAAAUACCAACAUUUUAUUAUUAUGCC ...................................((((.((......)).))))..(((((.....(((..........)))...(((((((......)))))))...))))). (-10.14 = -10.15 + 0.01)

| Location | 20,214,549 – 20,214,664 |
|---|---|
| Length | 115 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 80.44 |
| Shannon entropy | 0.43795 |
| G+C content | 0.31839 |
| Mean single sequence MFE | -21.90 |
| Consensus MFE | -14.48 |
| Energy contribution | -14.48 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.04 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
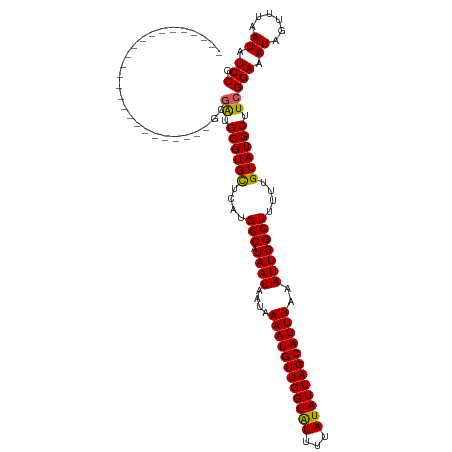
>dm3.chr3R 20214549 115 - 27905053 GGCAUAAUAAUAAAAUGUUGGUAUUUUAUAUUAGCAUUUAAAUUGGCUUUUUGUAUGCUUCGGAAAUAGUUUAAUAUCCGUAGAGGACCGUGCGUCUGAGUCGGGAGUCUGAAGU (((.((((....((((((((((((...))))))))))))..)))))))........((((((((............(((.....)))(((.((......)))))...)))))))) ( -25.70, z-score = -0.88, R) >droSim1.chr3R 20050708 115 - 27517382 GGCAUAAUAAUAAAAUGUUGGUAUUUUAUAUUAGCAUUUAAAUUGGCUUUUUGUAUGCUUCGGAAAUAGUUUAAUAUCCGUAGAGGACCGUGCGUCUGAGUCUCGAGUCUGAAGU (((.........((((((((((((...)))))))))))).....(((((..((((((((.((((.((......)).)))).)).....))))))...)))))....)))...... ( -23.80, z-score = -0.44, R) >droSec1.super_0 20553118 115 - 21120651 GGCAUAAUAAUAAAAUGUUGGUAUUUUAUAUUAGCAUUUAAAUUGGCUUUUUGUAUGCUUCGGAAAUAGUUUAAUAUCCGUAGAGGACCGUGCGUCUGAGUCUCAAGUCUGAAGU (((.........((((((((((((...)))))))))))).....(((((..((((((((.((((.((......)).)))).)).....))))))...)))))....)))...... ( -23.80, z-score = -0.70, R) >droYak2.chr3R 2542145 113 + 28832112 GGCAUAAUAAUAAAAUGUUGGUAUUUUAUAUUAGCAUUUAAAUUGGCUUUUUGUAUGCUUCGGAAAUAGUUUAAUAUCCGUAGAGGACCGUGAGUCGAAGUC-GAAGUC-GAGGU (((.........((((((((((((...))))))))))))...(((((((...((...((.((((.((......)).)))).))...))...))))))).)))-......-..... ( -24.70, z-score = -1.18, R) >droEre2.scaffold_4820 2624771 113 + 10470090 GGCAUAAUAAUAAAAUGUUGGUAUUUUAUAUUAGCAUUUAAAUUGGCUUUUUGUAUGCUUCGGAAAUAGUUUAAUAUCCGUAGAGGACCGUGAGUCUGAGUC-GGAGUC-GCACU (((.((((....((((((((((((...))))))))))))..)))))))....((...((.((((.((......)).)))).))...)).((((.((((...)-))).))-))... ( -24.20, z-score = -0.54, R) >droAna3.scaffold_13088 255142 110 - 569066 GGCAUAAUAAUAAAAUGUUGGUAUUUUAUAUUAGCAUUUAAAUUGGCUUUUUGUAUGCUUCGGAAAUAGUUUAAUAUCCGCAGAG----GAAUAGUCCAAGUCUGAGUCUAAGU- (((.........((((((((((((...)))))))))))).....(((((........((.((((.((......)).)))).)).(----((....))))))))...))).....- ( -20.90, z-score = -0.54, R) >dp4.chr2 12205452 115 + 30794189 GGCAUAAUAAUAAAAUGUUGGUAUUUUAUAUUAGCAUUUAAAUUGGCUUUUUGUAUGCUUCGGAAAUAGUUUAAUAUCCGUAGGGGCGAGGACUAUCAGAGUCAGAGUCUGUCUU ((((........((((((((((((...)))))))))))).....((((((((((...((.((((.((......)).)))).))..)))).((((.....)))))))))))))).. ( -25.90, z-score = -1.34, R) >droPer1.super_3 6055447 115 + 7375914 GGCAUAAUAAUAAAAUGUUGGUAUUUUAUAUUAGCAUUUAAAUUGGCUUUUUGUAUGCUUCGGAAAUAGUUUAAUAUCCGUAGGGGCGAGGACUAUCAGAGUCAGAGUCUGUCUU ((((........((((((((((((...)))))))))))).....((((((((((...((.((((.((......)).)))).))..)))).((((.....)))))))))))))).. ( -25.90, z-score = -1.34, R) >droWil1.scaffold_181108 216250 99 + 4707319 GGCAUAAUAAUAAAAUGUUGGUAUUUUAUAUUAGCAUUUAAAUUGGCUUUUUGUAUGCUUCGGAAAUAGUUUAAUAUCCGAAAAAGAGUCCCCGAUCUU---------------- ((..........((((((((((((...)))))))))))).....((((((((......((((((.((......)).)))))))))))))).))......---------------- ( -19.90, z-score = -1.36, R) >droMoj3.scaffold_6540 20565350 89 - 34148556 GGCAUAAUAAUAAAAUGUUGGUAUUUUAUAUUAGCAUUUAAAUUGGCUUUUUGUAUGCUUCGGAAAUAGUUUAAUAUCCGAAAAGUCUU-------------------------- .(((((.((((.((((((((((((...))))))))))))..))))........)))))((((((.((......)).)))))).......-------------------------- ( -17.10, z-score = -1.57, R) >droVir3.scaffold_13047 3559562 89 - 19223366 GGCAUAAUAAUAAAAUGUUGGUGUUUUAUAUUAGCAUUUAAAUUGGCUUUUUGUAUGCUUCGGAAAUAGUUUAAUAUCCGAAAAGUCUU-------------------------- .(((((.((((.((((((((((((...))))))))))))..))))........)))))((((((.((......)).)))))).......-------------------------- ( -17.10, z-score = -1.46, R) >droGri2.scaffold_14906 213847 89 - 14172833 GGCAUAAUAAUAAAACGUUGGUAUUUUAUAUUAGCAUUUAAAUUGGCUUUUUGUAUGCUUCGGAAAUAGUUUAAUAUCCGAAAAGUCUU-------------------------- .(((((..........((((((((...))))))))...((((.......)))))))))((((((.((......)).)))))).......-------------------------- ( -13.80, z-score = -0.31, R) >consensus GGCAUAAUAAUAAAAUGUUGGUAUUUUAUAUUAGCAUUUAAAUUGGCUUUUUGUAUGCUUCGGAAAUAGUUUAAUAUCCGUAGAGGACCGUGCGUCCGAGUC__GAGUC_GA__U ((((((.((((.((((((((((((...))))))))))))..))))........)))))).((((.((......)).))))................................... (-14.48 = -14.48 + 0.01)
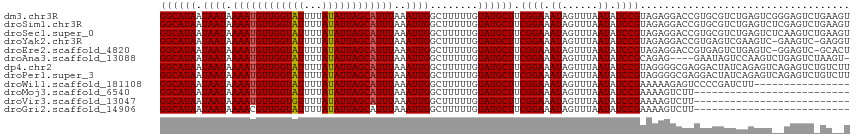


| Location | 20,214,584 – 20,214,679 |
|---|---|
| Length | 95 |
| Sequences | 10 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.14 |
| Shannon entropy | 0.28105 |
| G+C content | 0.33416 |
| Mean single sequence MFE | -23.45 |
| Consensus MFE | -21.50 |
| Energy contribution | -21.36 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.859308 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 20214584 95 - 27905053 -------------------------GCGAUGCGUGCUCAUGGCAUAAUAAUAAAAUGUUGGUAUUUUAUAUUAGCAUUUAAAUUGGCUUUUUGUAUGCUUCGGAAAUAGUUUAAUAUCCG -------------------------..((.((((((....(((.((((....((((((((((((...))))))))))))..)))))))....)))))).))(((.((......)).))). ( -23.20, z-score = -1.94, R) >droSim1.chr3R 20050743 95 - 27517382 -------------------------GCGAUGCGUGCUCAUGGCAUAAUAAUAAAAUGUUGGUAUUUUAUAUUAGCAUUUAAAUUGGCUUUUUGUAUGCUUCGGAAAUAGUUUAAUAUCCG -------------------------..((.((((((....(((.((((....((((((((((((...))))))))))))..)))))))....)))))).))(((.((......)).))). ( -23.20, z-score = -1.94, R) >droSec1.super_0 20553153 95 - 21120651 -------------------------GCGAUGCGUGCUCAUGGCAUAAUAAUAAAAUGUUGGUAUUUUAUAUUAGCAUUUAAAUUGGCUUUUUGUAUGCUUCGGAAAUAGUUUAAUAUCCG -------------------------..((.((((((....(((.((((....((((((((((((...))))))))))))..)))))))....)))))).))(((.((......)).))). ( -23.20, z-score = -1.94, R) >droYak2.chr3R 2542178 95 + 28832112 -------------------------GCGAUGCGUGCUCAUGGCAUAAUAAUAAAAUGUUGGUAUUUUAUAUUAGCAUUUAAAUUGGCUUUUUGUAUGCUUCGGAAAUAGUUUAAUAUCCG -------------------------..((.((((((....(((.((((....((((((((((((...))))))))))))..)))))))....)))))).))(((.((......)).))). ( -23.20, z-score = -1.94, R) >droEre2.scaffold_4820 2624804 95 + 10470090 -------------------------GCGAUGCGUGCUCAUGGCAUAAUAAUAAAAUGUUGGUAUUUUAUAUUAGCAUUUAAAUUGGCUUUUUGUAUGCUUCGGAAAUAGUUUAAUAUCCG -------------------------..((.((((((....(((.((((....((((((((((((...))))))))))))..)))))))....)))))).))(((.((......)).))). ( -23.20, z-score = -1.94, R) >droAna3.scaffold_13088 255172 106 - 569066 --------------GGAAUCUGUAGGGGAUGCGUGCUCUUGGCAUAAUAAUAAAAUGUUGGUAUUUUAUAUUAGCAUUUAAAUUGGCUUUUUGUAUGCUUCGGAAAUAGUUUAAUAUCCG --------------(((((((.....))))((((((....(((.((((....((((((((((((...))))))))))))..)))))))....))))))..................))). ( -23.10, z-score = -1.16, R) >dp4.chr2 12205487 106 + 30794189 --------------GAGAGGGAGAGGGGGUGCGUGCUCUUGGCAUAAUAAUAAAAUGUUGGUAUUUUAUAUUAGCAUUUAAAUUGGCUUUUUGUAUGCUUCGGAAAUAGUUUAAUAUCCG --------------.............((.((((((....(((.((((....((((((((((((...))))))))))))..)))))))....)))))).))(((.((......)).))). ( -22.00, z-score = -0.43, R) >droPer1.super_3 6055482 106 + 7375914 --------------GAGAGGGAGAGGGGGUGCGUGCUCUUGGCAUAAUAAUAAAAUGUUGGUAUUUUAUAUUAGCAUUUAAAUUGGCUUUUUGUAUGCUUCGGAAAUAGUUUAAUAUCCG --------------.............((.((((((....(((.((((....((((((((((((...))))))))))))..)))))))....)))))).))(((.((......)).))). ( -22.00, z-score = -0.43, R) >droWil1.scaffold_181108 216269 112 + 4707319 -------AGACCUCCCAAUGUAUGUGGCA-GCGUGCGCCUGGCAUAAUAAUAAAAUGUUGGUAUUUUAUAUUAGCAUUUAAAUUGGCUUUUUGUAUGCUUCGGAAAUAGUUUAAUAUCCG -------.......(((.......))).(-(((((((...(((.((((....((((((((((((...))))))))))))..)))))))...)))))))).((((.((......)).)))) ( -23.20, z-score = 0.14, R) >droVir3.scaffold_13047 3559571 120 - 19223366 GUGCUUGUAUGCGUGUGCCUUAGUGUGAAUGCGUGUGGCUGGCAUAAUAAUAAAAUGUUGGUGUUUUAUAUUAGCAUUUAAAUUGGCUUUUUGUAUGCUUCGGAAAUAGUUUAAUAUCCG ......(((((((...(((......((((((((((((((..((((.........))))..))....)))))..)))))))....)))....)))))))..((((.((......)).)))) ( -28.20, z-score = -0.69, R) >consensus _________________________GCGAUGCGUGCUCAUGGCAUAAUAAUAAAAUGUUGGUAUUUUAUAUUAGCAUUUAAAUUGGCUUUUUGUAUGCUUCGGAAAUAGUUUAAUAUCCG ...........................((.((((((....(((.((((....((((((((((((...))))))))))))..)))))))....)))))).))(((.((......)).))). (-21.50 = -21.36 + -0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:40:22 2011