
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,198,110 – 20,198,226 |
| Length | 116 |
| Max. P | 0.579002 |

| Location | 20,198,110 – 20,198,226 |
|---|---|
| Length | 116 |
| Sequences | 9 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 79.51 |
| Shannon entropy | 0.41577 |
| G+C content | 0.54885 |
| Mean single sequence MFE | -33.80 |
| Consensus MFE | -19.12 |
| Energy contribution | -19.36 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.578347 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
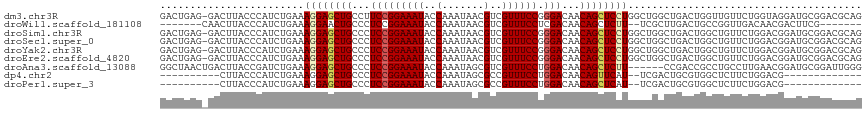
>dm3.chr3R 20198110 116 + 27905053 GACUGAG-GACUUACCCAUCUGAAAGGAGCUGCCUUCCGGAAAUACCAAAUAACGUCGUUUCCGGGACAACAGCUCCUGGCUGGCUGACUGGUUGUUCUGGUAGGAUGCGGACGCAG .(((.((-(((..(((..((.(..((((((((..((((((((((((........)).))))))))))...))))))))..).....))..))).))))))))....(((....))). ( -40.10, z-score = -1.63, R) >droWil1.scaffold_181108 199615 101 - 4707319 -------CAACUUACCCAUCUGAAAGGAACUGCCCUCCGGAAAUACCAAAUAACGUCGUUUCCUCGACAACAGCUCUU--UCGCUUGACUGCCGGUUGACAACGACUUCG------- -------...........((((..(((......))).)))).............((((((...(((((..(((.((..--......)))))...))))).))))))....------- ( -19.90, z-score = -1.21, R) >droSim1.chr3R 20032992 116 + 27517382 GACUGAG-GACUUACCCAUCUGAAAGGAGCUGCCCUCCGGAAAUACCAAAUAACGUCGUUUCCGGGACAACAGCUCCUGGCUGGCUGACUGGCUGUUCUGGACGGAUGCGGACGCAG .......-......((((((((..((((((((...(((((((((((........)).)))))))))....))))))))(((..(....)..)))........)))))).))...... ( -41.50, z-score = -2.12, R) >droSec1.super_0 20536896 116 + 21120651 GACUGAG-GACUUACCCAUCUGAAAGGAGCUGCCCUCCGGAAAUACCAAAUAACGUCGUUUCCGGGACAACAGCUCCUGGCUGGCUGACUGGCUGUUCUGGACGGAUGCGGACGCAG .......-......((((((((..((((((((...(((((((((((........)).)))))))))....))))))))(((..(....)..)))........)))))).))...... ( -41.50, z-score = -2.12, R) >droYak2.chr3R 2525299 116 - 28832112 GACUGAG-GACUUACCCAUCUGAAAGGAGCUGCCCUCCGGAAAUACCAAAUAACGUCGUUUCCGGGACAACAGCUCCUGGCUGGCUGACUGGCUGUUCUGGACGGAUGCGGACGCAG .......-......((((((((..((((((((...(((((((((((........)).)))))))))....))))))))(((..(....)..)))........)))))).))...... ( -41.50, z-score = -2.12, R) >droEre2.scaffold_4820 2608498 116 - 10470090 GACUGAG-GACUUACCCAUCUGAAAGGAGCUGCCCUCCGGAAAUACCAAAUAACGUCGUUUCCGGGACAACAGCUCCUGGCUGGCUGACUGGCUGUUCUGGACGGAUGCGGACGCAG .......-......((((((((..((((((((...(((((((((((........)).)))))))))....))))))))(((..(....)..)))........)))))).))...... ( -41.50, z-score = -2.12, R) >droAna3.scaffold_13088 238282 111 + 569066 GGCUAACUGACUUACCGAUCUGAAAGGAGCUGCCCUCCGGAAAUACCAAAUAGCGUCGUUUCCUGGACAACAGCUCUU------CCGACCGCCUGCCUUGAACGGAUGCGGAUUGGG ..............((((((((...(((((((...((((((((((((.....).)).)))))).)))...)))))))(------(((..((.......))..))))..)))))))). ( -33.80, z-score = -1.54, R) >dp4.chr2 12184087 92 - 30794189 ----------CUUACCCAUCUGAAAGGAGCUGCCCUCCGGAAAUACCAAAUAGCGCCGUUUCCUGGACAACAGUUCAU--UCGACUGCGUGGCUCUUCUGGACG------------- ----------.....(((...((..(((((..(..(((((((((..(.......)..)))))).)))...(((((...--..))))).)..))))))))))...------------- ( -22.70, z-score = -0.44, R) >droPer1.super_3 6035751 92 - 7375914 ----------CUUACCCAUCUGAAAGGAGCUGCCCUCCGGAAAUACCAAAUAGCGCCGUUUCCUGGACAACAGCUCAU--UCGACUGCGUGGCUCUUCUGGACG------------- ----------.((((.((..((((..((((((...(((((((((..(.......)..)))))).)))...)))))).)--)))..)).))))............------------- ( -21.70, z-score = -0.07, R) >consensus GACUGAG_GACUUACCCAUCUGAAAGGAGCUGCCCUCCGGAAAUACCAAAUAACGUCGUUUCCGGGACAACAGCUCCUGGCUGGCUGACUGGCUGUUCUGGACGGAUGCGGACGCAG ........................((((((((...(((((((((..(.......)..)))))).)))...))))))))....................................... (-19.12 = -19.36 + 0.24)

| Location | 20,198,110 – 20,198,226 |
|---|---|
| Length | 116 |
| Sequences | 9 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 79.51 |
| Shannon entropy | 0.41577 |
| G+C content | 0.54885 |
| Mean single sequence MFE | -35.12 |
| Consensus MFE | -22.13 |
| Energy contribution | -22.67 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.579002 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
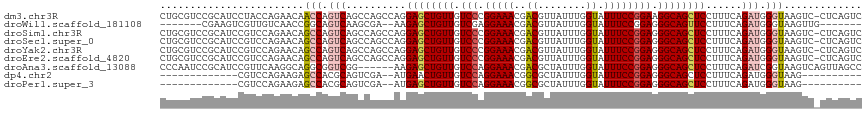
>dm3.chr3R 20198110 116 - 27905053 CUGCGUCCGCAUCCUACCAGAACAACCAGUCAGCCAGCCAGGAGCUGUUGUCCCGGAAACGACGUUAUUUGGUAUUUCCGGAAGGCAGCUCCUUUCAGAUGGGUAAGUC-CUCAGUC (((.(..(..((((.....((........)).....(..((((((((((...(((((((...((.....))...)))))))..))))))))))..)....))))..)..-).))).. ( -33.70, z-score = -0.68, R) >droWil1.scaffold_181108 199615 101 + 4707319 -------CGAAGUCGUUGUCAACCGGCAGUCAAGCGA--AAGAGCUGUUGUCGAGGAAACGACGUUAUUUGGUAUUUCCGGAGGGCAGUUCCUUUCAGAUGGGUAAGUUG------- -------....(((((((((....))))).)....((--((((((((((.((..(((((...((.....))...))))).)).))))))).))))).)))..........------- ( -26.90, z-score = -0.39, R) >droSim1.chr3R 20032992 116 - 27517382 CUGCGUCCGCAUCCGUCCAGAACAGCCAGUCAGCCAGCCAGGAGCUGUUGUCCCGGAAACGACGUUAUUUGGUAUUUCCGGAGGGCAGCUCCUUUCAGAUGGGUAAGUC-CUCAGUC (((.(..(..(((((((..(((..((......)).....((((((((((.(((.(((((...((.....))...)))))))).))))))))))))).)))))))..)..-).))).. ( -40.90, z-score = -2.11, R) >droSec1.super_0 20536896 116 - 21120651 CUGCGUCCGCAUCCGUCCAGAACAGCCAGUCAGCCAGCCAGGAGCUGUUGUCCCGGAAACGACGUUAUUUGGUAUUUCCGGAGGGCAGCUCCUUUCAGAUGGGUAAGUC-CUCAGUC (((.(..(..(((((((..(((..((......)).....((((((((((.(((.(((((...((.....))...)))))))).))))))))))))).)))))))..)..-).))).. ( -40.90, z-score = -2.11, R) >droYak2.chr3R 2525299 116 + 28832112 CUGCGUCCGCAUCCGUCCAGAACAGCCAGUCAGCCAGCCAGGAGCUGUUGUCCCGGAAACGACGUUAUUUGGUAUUUCCGGAGGGCAGCUCCUUUCAGAUGGGUAAGUC-CUCAGUC (((.(..(..(((((((..(((..((......)).....((((((((((.(((.(((((...((.....))...)))))))).))))))))))))).)))))))..)..-).))).. ( -40.90, z-score = -2.11, R) >droEre2.scaffold_4820 2608498 116 + 10470090 CUGCGUCCGCAUCCGUCCAGAACAGCCAGUCAGCCAGCCAGGAGCUGUUGUCCCGGAAACGACGUUAUUUGGUAUUUCCGGAGGGCAGCUCCUUUCAGAUGGGUAAGUC-CUCAGUC (((.(..(..(((((((..(((..((......)).....((((((((((.(((.(((((...((.....))...)))))))).))))))))))))).)))))))..)..-).))).. ( -40.90, z-score = -2.11, R) >droAna3.scaffold_13088 238282 111 - 569066 CCCAAUCCGCAUCCGUUCAAGGCAGGCGGUCGG------AAGAGCUGUUGUCCAGGAAACGACGCUAUUUGGUAUUUCCGGAGGGCAGCUCCUUUCAGAUCGGUAAGUCAGUUAGCC .((...((((..((......))...))))..((------((((((((((.(((.(((((..((........)).)))))))).))))))).))))).....)).............. ( -36.70, z-score = -1.22, R) >dp4.chr2 12184087 92 + 30794189 -------------CGUCCAGAAGAGCCACGCAGUCGA--AUGAACUGUUGUCCAGGAAACGGCGCUAUUUGGUAUUUCCGGAGGGCAGCUCCUUUCAGAUGGGUAAG---------- -------------((((..((((.(((..(((((...--....)))))......(....))))(((.(((((.....))))).))).....))))..))))......---------- ( -25.10, z-score = 0.29, R) >droPer1.super_3 6035751 92 + 7375914 -------------CGUCCAGAAGAGCCACGCAGUCGA--AUGAGCUGUUGUCCAGGAAACGGCGCUAUUUGGUAUUUCCGGAGGGCAGCUCCUUUCAGAUGGGUAAG---------- -------------...........(((.....(((((--(.((((((((.(((.(((((..((........)).)))))))).))))))))..))).))).)))...---------- ( -30.10, z-score = -0.99, R) >consensus CUGCGUCCGCAUCCGUCCAGAACAGCCAGUCAGCCAGCCAGGAGCUGUUGUCCCGGAAACGACGUUAUUUGGUAUUUCCGGAGGGCAGCUCCUUUCAGAUGGGUAAGUC_CUCAGUC ........................(((.(((..........((((((((.(((.(((((..((........)).)))))))).))))))))......))).)))............. (-22.13 = -22.67 + 0.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:40:18 2011