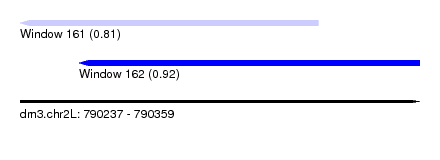
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 790,237 – 790,359 |
| Length | 122 |
| Max. P | 0.917607 |

| Location | 790,237 – 790,328 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 69.06 |
| Shannon entropy | 0.58280 |
| G+C content | 0.42889 |
| Mean single sequence MFE | -16.84 |
| Consensus MFE | -8.23 |
| Energy contribution | -9.14 |
| Covariance contribution | 0.91 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.810197 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
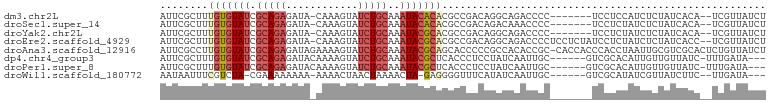
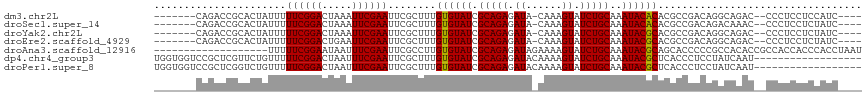
>dm3.chr2L 790237 91 - 23011544 AUUCGCUUUGUGUAUCGCAGAGAUA-CAAAGUAUCUGCAAAUACACACGCCGACAGGCAGACCCC-------UCCUCCAUCUCUAUCACA--UCGUUAUCU ........(((((((.(((((.((.-....)).)))))..))))))).(((....))).......-------..................--......... ( -18.00, z-score = -1.88, R) >droSec1.super_14 760261 91 - 2068291 AUUCGCUUUGUGUAUCGCAGAGAUA-CAAAGUAUCUGCAAAUACACACGCCGACAGACAAACCCC-------UCCUCUAUCUCUAUCACA--UCGUUAUCU ..(((...(((((((.(((((.((.-....)).)))))..)))))))...)))............-------..................--......... ( -14.80, z-score = -1.78, R) >droYak2.chr2L 772688 91 - 22324452 AUUCGCUUUGUGUAUCGCAGAGAUA-CAAAGUAUCUGCAAAUACGCACGCCGACAGGCAGACCCC-------UCCUCUAUCUCUAUCACA--UCGUUAUCU ........(((((((.(((((.((.-....)).)))))..))))))).(((....))).......-------..................--......... ( -17.60, z-score = -1.06, R) >droEre2.scaffold_4929 844114 98 - 26641161 AUUCGCUUUGUGUAUCGCAGAGAUA-CAAAGUAUCUGCAAAUACGCACGCCGACAGGCAGACCCCUCCUCUAUCCUCUAUCUCUAUCACC--UCGUUAUCU ........(((((((.(((((.((.-....)).)))))..))))))).(((....)))................................--......... ( -17.60, z-score = -1.07, R) >droAna3.scaffold_12916 1754450 100 - 16180835 AUUCGCCUUGUGUAUCGCAGAGAUAGAAAAGUAUCUGCAAAUACGCAGCACCCCCGCCACACCGC-CACCACCCACCUAAUUGCGUCGCACUCUGUUAUCU ....((..(((((((.(((((.((......)).)))))..)))))))))..............((-.((((..........)).)).))............ ( -16.20, z-score = -0.33, R) >dp4.chr4_group3 2748574 91 + 11692001 AUUCGCUUUGUGUAUCGCAGAGAUACAAAAGUAUCUGCAAAUACGCUCACCCUCCUAUCAAUUGC------GUCGCACAUUGUUGUUAUC-UUUGAUA--- ....((..(((((..(((((((((((....))))))((......))...............))))------)..)))))..)).......-.......--- ( -20.90, z-score = -2.20, R) >droPer1.super_8 3838745 91 + 3966273 AUUCGCUUUGUGUAUCGCAGAGAUACAAAAGUAUCUGCAAAUACGCUCACCCUCCUAUCAAUUGC------GUCGCACAUUGUUGUUAUC-UUUGAUA--- ....((..(((((..(((((((((((....))))))((......))...............))))------)..)))))..)).......-.......--- ( -20.90, z-score = -2.20, R) >droWil1.scaffold_180772 6465690 87 + 8906247 AAUAAUUUCGUCUA-CGAAAAAAAA-AAAACUAACUAAAACUA-GAGGGGUUUCAUAUCAAUUGC------GUCGCAUAUCGUUAUCUUC--UUGAUA--- .....(((((....-))))).....-.(((((..(((....))-)...)))))..((((((..((------(........))).......--))))))--- ( -8.70, z-score = 0.35, R) >consensus AUUCGCUUUGUGUAUCGCAGAGAUA_AAAAGUAUCUGCAAAUACGCACGCCGACAGACAAACCCC_______UCCCCUAUCUCUAUCACA__UCGUUAUCU ........(((((((.(((((.((......)).)))))..)))))))...................................................... ( -8.23 = -9.14 + 0.91)
| Location | 790,255 – 790,359 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 74.65 |
| Shannon entropy | 0.46137 |
| G+C content | 0.45859 |
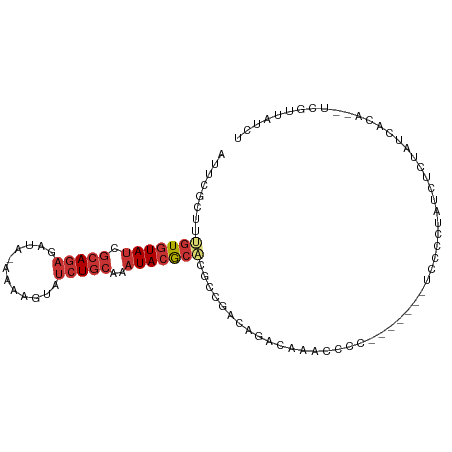
| Mean single sequence MFE | -21.16 |
| Consensus MFE | -15.43 |
| Energy contribution | -15.23 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.917607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
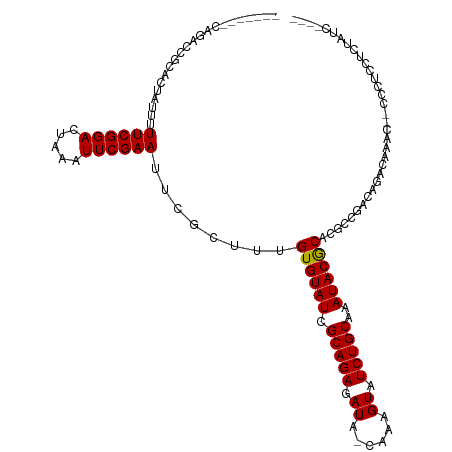
>dm3.chr2L 790255 104 - 23011544 -------CAGACCGCACUAUUUUUCGGACUAAAUUCGAAUUCGCUUUGUGUAUCGCAGAGAUA-CAAAGUAUCUGCAAAUACACACGCCGACAGGCAGAC--CCCUCCUCCAUC---- -------......((.......((((((.....))))))...))..(((((((.(((((.((.-....)).)))))..))))))).(((....)))....--............---- ( -21.40, z-score = -1.50, R) >droSec1.super_14 760279 104 - 2068291 -------CAGACCGCACUAUUUUUCGGACUAAAUUCGAAUUCGCUUUGUGUAUCGCAGAGAUA-CAAAGUAUCUGCAAAUACACACGCCGACAGACAAAC--CCCUCCUCUAUC---- -------.(((...........((((((.....)))))).(((...(((((((.(((((.((.-....)).)))))..)))))))...))).........--......)))...---- ( -18.60, z-score = -1.33, R) >droYak2.chr2L 772706 104 - 22324452 -------CAGACCGCACUAUUUUUCGGACUAAAUUCGAAUUCGCUUUGUGUAUCGCAGAGAUA-CAAAGUAUCUGCAAAUACGCACGCCGACAGGCAGAC--CCCUCCUCUAUC---- -------.(((...........((((((.....)))))).((....(((((((.(((((.((.-....)).)))))..))))))).(((....))).)).--......)))...---- ( -21.30, z-score = -0.86, R) >droEre2.scaffold_4929 844139 104 - 26641161 -------CAGACCGCACUAUUUUUCGGACUGAAUUCGAAUUCGCUUUGUGUAUCGCAGAGAUA-CAAAGUAUCUGCAAAUACGCACGCCGACAGGCAGAC--CCCUCCUCUAUC---- -------(((.(((..........))).)))...............(((((((.(((((.((.-....)).)))))..))))))).(((....)))....--............---- ( -21.80, z-score = -0.56, R) >droAna3.scaffold_12916 1754470 99 - 16180835 -------------------UUUUUCGGAAUAAUUUCGAAUUCGCCUUGUGUAUCGCAGAGAUAGAAAAGUAUCUGCAAAUACGCAGCACCCCCGCCACACCGCCACCACCCACCUAAU -------------------...((((((.....))))))...((..(((((((.(((((.((......)).)))))..)))))))))............................... ( -17.90, z-score = -1.87, R) >dp4.chr4_group3 2748603 100 + 11692001 UGGUGGUCCGCUCGUUCUGUUUUUCGGACUAAUUUCGAAUUCGCUUUGUGUAUCGCAGAGAUACAAAAGUAUCUGCAAAUACGCUCACCCUCCUAUCAAU------------------ .(((((((((..............)))))..................((((((.(((((..(((....))))))))..)))))).))))...........------------------ ( -22.54, z-score = -1.39, R) >droPer1.super_8 3838774 100 + 3966273 UGGUGGUCCGCUCGGUCUGUUUUUCGGACUAAUUUCGAAUUCGCUUUGUGUAUCGCAGAGAUACAAAAGUAUCUGCAAAUACGCUCACCCUCCUAUCAAU------------------ .(((((...((..((((((.....))))))......(....)))...((((((.(((((..(((....))))))))..)))))))))))...........------------------ ( -24.60, z-score = -1.65, R) >consensus _______CAGACCGCACUAUUUUUCGGACUAAAUUCGAAUUCGCUUUGUGUAUCGCAGAGAUA_CAAAGUAUCUGCAAAUACGCACGCCGACAGACAAAC__CCCUCCUCUAUC____ ......................((((((.....))))))........((((((.(((((.((......)).)))))..)))))).................................. (-15.43 = -15.23 + -0.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:06:54 2011