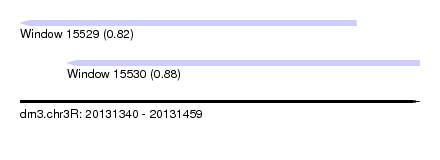
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,131,340 – 20,131,459 |
| Length | 119 |
| Max. P | 0.882690 |

| Location | 20,131,340 – 20,131,440 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 63.07 |
| Shannon entropy | 0.64861 |
| G+C content | 0.41788 |
| Mean single sequence MFE | -18.31 |
| Consensus MFE | -8.45 |
| Energy contribution | -9.38 |
| Covariance contribution | 0.93 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.823380 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
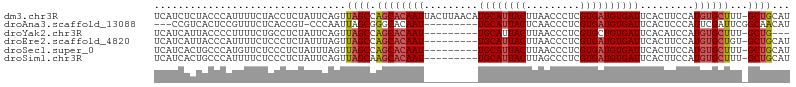
>dm3.chr3R 20131340 100 - 27905053 --------ACCUCUAUUCAGUUAGCCAGCACAAUUACUUAACAUGCAUUACUUAACCCUCGUGAUGUGAUUCACUUCCAUGUGCUUU-GCUGCAUGGCACGAUUGUCCA------------ --------.........(((((.((((((((((...........(((((((.........)))))))....(((......)))..))-).))).))))..)))))....------------ ( -19.80, z-score = -0.81, R) >dp4.chrXL_group1e 2563236 118 - 12523060 UACUCUAGAUCGAUAUAUAUCUGAUUAAUACAGAUAUACAA-AAAUAAUGCUUAUCAUAUAUAAAAUCAUAUACAUAUAUAAGAUCA-UCUGC-UUAUGUAUUCCUAUAGGACCUUCUCUG .....(((((.(((.((((((((.......))))))))...-........(((((.((((.((........)).)))))))))))))-))))(-(((((......)))))).......... ( -15.70, z-score = -0.06, R) >droAna3.scaffold_13088 169468 89 - 569066 -----------CCGUCCCAAUUAGCGGGCACAAU---------UGCAUUACUCAACCCUCGUGAUGUGAUUCACUCCCAUUCGAUUCGGCAACAUUUCACGUUUGUUUA------------ -----------.......(((.((((((...(((---------..((((((.........))))))..)))....)))....((...(....)...))..))).)))..------------ ( -16.50, z-score = -0.48, R) >droYak2.chr3R 2457734 78 + 28832112 --------GCCUCUAUUCAGUUAGCCAGCACAAU---------UGCAUUACUUAACCCUCGUGCUGUGAUUCACAUCCAUGUGCUUU-GCUGUCCA------------------------- --------.............((((.((((((..---------.((((............))))((((...))))....))))))..-))))....------------------------- ( -14.10, z-score = -0.64, R) >droEre2.scaffold_4820 2540551 91 + 10470090 --------CCCUCUAUUUAGUUAGCCAGCACAAU---------UGCAUUACUUAACCCUCGUGAUGUGAUUCACUUCCAUGUGCUGU-GCUGCAUUUCACGUUUGUUCA------------ --------...........((.((((((((((((---------..((((((.........))))))..)).........))))))).-)))))................------------ ( -22.50, z-score = -3.08, R) >droSec1.super_0 20469873 91 - 21120651 --------CCCUCUAUUUAGUUAGCCAGCACAAU---------UGCAUUACUUAACCCUCGUGAUGUGAUUCACUUCCAUGUGCUUU-GCUGCAUUGCACGAUUGUCCA------------ --------.........((((((((.((((((((---------..((((((.........))))))..)).........))))))..-)))).))))............------------ ( -18.00, z-score = -1.28, R) >droSim1.chr3R 19965941 91 - 27517382 --------CCCUCUAUUCAGUUAGCAAGCACAAU---------UGCAUUACUUAGCCCUCGUGAUGUGAUUCACUUCCAUGUGCUUU-GCUGCAUUGCACGAUUGUCCA------------ --------.........(((((((((((((((((---------..((((((.........))))))..)).........))))).))-)))).))))............------------ ( -21.60, z-score = -1.65, R) >consensus ________CCCUCUAUUCAGUUAGCCAGCACAAU_________UGCAUUACUUAACCCUCGUGAUGUGAUUCACUUCCAUGUGCUUU_GCUGCAUUGCACGAUUGUCCA____________ .....................((((.((((((...........((((((((.........))))))))...........))))))...))))............................. ( -8.45 = -9.38 + 0.93)



| Location | 20,131,354 – 20,131,459 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 82.21 |
| Shannon entropy | 0.32227 |
| G+C content | 0.43965 |
| Mean single sequence MFE | -18.25 |
| Consensus MFE | -12.39 |
| Energy contribution | -12.95 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.882690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 20131354 105 - 27905053 UCAUCUCUACCCAUUUUCUACCUCUAUUCAGUUAGCCAGCACAAUUACUUAACAUGCAUUACUUAACCCUCGUGAUGUGAUUCACUUCCAUGUGCUUU-GCUGCAU ..............................((.(((.((((((............(((((((.........)))))))((......))..))))))..-))))).. ( -16.50, z-score = -2.05, R) >droAna3.scaffold_13088 169482 93 - 569066 ---CCGUCACUCCGUUUCUCACCGU-CCCAAUUAGCGGGCACAAU---------UGCAUUACUCAACCCUCGUGAUGUGAUUCACUCCCAUUCGAUUCGGCAACAU ---........(((..((.....((-(((.......))).))(((---------..((((((.........))))))..)))...........))..)))...... ( -19.00, z-score = -1.35, R) >droYak2.chr3R 2457738 93 + 28832112 UCAUCAUUACCCCUUUUCUGCCUCUAUUCAGUUAGCCAGCACAAU---------UGCAUUACUUAACCCUCGUGCUGUGAUUCACAUCCAUGUGCUUU-GCUG--- ................................((((.((((((..---------.((((............))))((((...))))....))))))..-))))--- ( -13.50, z-score = -0.98, R) >droEre2.scaffold_4820 2540565 96 + 10470090 UCAUCAUUACCCAUUUUCUCCCUCUAUUUAGUUAGCCAGCACAAU---------UGCAUUACUUAACCCUCGUGAUGUGAUUCACUUCCAUGUGCUGU-GCUGCAU ..............................((.((((((((((((---------..((((((.........))))))..)).........))))))).-))))).. ( -22.50, z-score = -3.98, R) >droSec1.super_0 20469887 96 - 21120651 UCAUCACUGCCCAUGUUCUCCCUCUAUUUAGUUAGCCAGCACAAU---------UGCAUUACUUAACCCUCGUGAUGUGAUUCACUUCCAUGUGCUUU-GCUGCAU ..............................((.(((.((((((((---------..((((((.........))))))..)).........))))))..-))))).. ( -17.90, z-score = -1.61, R) >droSim1.chr3R 19965955 96 - 27517382 UCAUCACUGCCCAUUUUCUCCCUCUAUUCAGUUAGCAAGCACAAU---------UGCAUUACUUAGCCCUCGUGAUGUGAUUCACUUCCAUGUGCUUU-GCUGCAU ..............................((.((((((((((((---------..((((((.........))))))..)).........))))).))-))))).. ( -20.10, z-score = -2.12, R) >consensus UCAUCACUACCCAUUUUCUCCCUCUAUUCAGUUAGCCAGCACAAU_________UGCAUUACUUAACCCUCGUGAUGUGAUUCACUUCCAUGUGCUUU_GCUGCAU ................................((((.((((((...........((((((((.........))))))))...........))))))...))))... (-12.39 = -12.95 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:40:15 2011