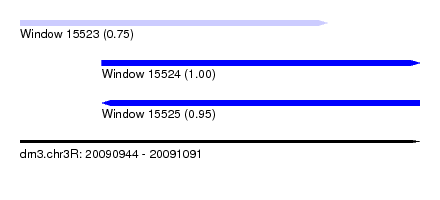
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,090,944 – 20,091,091 |
| Length | 147 |
| Max. P | 0.995143 |

| Location | 20,090,944 – 20,091,057 |
|---|---|
| Length | 113 |
| Sequences | 13 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 84.32 |
| Shannon entropy | 0.34177 |
| G+C content | 0.54358 |
| Mean single sequence MFE | -40.92 |
| Consensus MFE | -29.70 |
| Energy contribution | -29.33 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.748312 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 20090944 113 + 27905053 -----UUGGUUUUAUAGCUUUUUUG--GCGACGAUUCUGCAC-AGUCAGGUUGGCCGAGCGGUCUAAGGCGCCAGAUUUAAGCUCUGGUUCUCGAGAGGGAGCGUGGGUUCGAACCCCACA -----..........(((((.((((--(((......((((.(-.(((.....))).).)))).......)))))))...)))))...((((((....))))))(((((.......))))). ( -40.92, z-score = -1.49, R) >droSim1.chr3R 19923637 113 + 27517382 -----UCGCUUUUAUAGCUCUUUUG--GCGGCGAUUCUGCAC-AGUCAGGUUGGCCGAGCGGUCUAAGGCGCCAGAUUUAAGCUCUGGUUCUCGAGAGGGAGCGUGGGUUCGAACCCCACA -----..........((((..((((--(((.(....((((.(-.(((.....))).).)))).....).)))))))....))))...((((((....))))))(((((.......))))). ( -41.50, z-score = -0.85, R) >droSec1.super_0 20423388 113 + 21120651 -----UCGCUUUUAUAGCUCUUUUG--UCGGCGAUUCUGCAC-AGUCAGGUUGGCCGAGCGGUCUAAGGCGCCAGAUUUAAGCUCUGGUUCUCGAGAGGGAGCGUGGGUUCGAACCCCACA -----...........(((((((((--(((((.(..(((...-...)))..).))))).)))(((..((.((((((.......)))))).))..)))))))))(((((.......))))). ( -39.50, z-score = -0.64, R) >droYak2.chr3R 2416176 113 - 28832112 -----UCGCUUAAACAGUUCUUUUG--UCGGCGAUUCUGCAC-AGUCAGGUUGGCCGAGCGGUCUAAGGCGCCAGAUUUAAGCUCUGGUUCCCGAGAGGGAGCGUGGGUUCGAACCCCACA -----.........(((..(((..(--((((((...((((.(-.(((.....))).).)))).......)))).)))..)))..)))((((((....))))))(((((.......))))). ( -43.10, z-score = -1.71, R) >droEre2.scaffold_4820 2498284 113 - 10470090 -----UCGCUCAAAUAGCUCUUCUG--UCGUCGAUGCUGCAC-AGUCAGGUUGGCCGAGCGGUCUAAGGCGCCAGAUUUAAGCUUUGGUUCUCGAGAGGGAGCGUGGGUUCGAACCCCACA -----.....(((..((((..((((--.((((...(((((.(-.(((.....))).).)))))....)))).))))....)))))))((((((....))))))(((((.......))))). ( -41.00, z-score = -0.98, R) >droAna3.scaffold_13340 1291715 115 - 23697760 -----UUGCUUCUAUAGCACAUUUUGU-CGACGUCGCCCAACAAGUCAGGUUGGCCGAGCGGUCUAAGGCGCCAGAUUUAAGCUCUGGUUCUCGAAAGAGAGCGUGGGUUCGAACCCCACA -----.((((.....))))........-.(((((((.(((((.......))))).))).).)))....((((((((.......))))))((((....))))))(((((.......))))). ( -38.60, z-score = -1.38, R) >dp4.chr2 14106006 113 + 30794189 -------GCUUAUAUAGCUCGUUUAGA-CAACGACGCCCGACGAGUCAGGUUGGCCGAGCGGUCUAAGGCGCCAGAUUUAAGCUCUGGUUCUCGAGAGAGAGCGUGGGUUCGAACCCCACA -------(((((.((.((.(.((((((-(..(..((.(((((.......))))).)).)..)))))))).))...)).)))))....((((((....))))))(((((.......))))). ( -41.10, z-score = -1.58, R) >droPer1.super_0 5486811 113 - 11822988 -------GCUCAUAUAGCUCGUUUAGA-UAACGACGCCCGACGAGUCAGGUUGGCCGAGCGGUCUAAGGCGCCAGAUUUAAGCUCUGGUUCUCGAGAGAGAGCGUGGGUUCGAACCCCACA -------((((.....(((((((....-.))))).))(((((.......)))))..))))........((((((((.......))))))((((....))))))(((((.......))))). ( -40.50, z-score = -1.35, R) >droWil1.scaffold_181089 3151105 120 + 12369635 AUGAAAUAUUCAUGUAGCACAUUCGGAGCACAGGCAGACA-CAAGUCAGGUUGGCCGAGCGGUCUAAGGCGCCAGAUUUAAGCUCUGGUUCUCGAGAGAGAGCGUGGGUUCGAACCCCACA ((((.....))))..........((((((...(((.(((.-(......)))).)))(.(((........))))........))))))((((((....))))))(((((.......))))). ( -38.60, z-score = -1.31, R) >droVir3.scaffold_13047 10926503 115 + 19223366 -----UUGCUUAUGUAGCACGUUUAGAGC-UACUCGCCCAACAAGUCAGGUUGGCCGAGCGGUCUAAGGCGCCAGAUUUAAGCUCUGGUUCUCGAGAGAGAGCGUGGGUUCGAACCCCACA -----..(((((.((.((.(.((((((.(-..((((.(((((.......))))).))))..)))))))).))...)).)))))....((((((....))))))(((((.......))))). ( -44.50, z-score = -2.54, R) >droMoj3.scaffold_6540 3299129 115 - 34148556 -----CUGCUUAUGUAGCACGUUUAGA-CAGACCCGUUCAACAAGUCAGGUUGGCCGAGCGGUCUAAGGCGCCAGAUUUAAGCUCUGGUUCUCGAGAGAGAGCGUGGGUUCGAACCCCACA -----..(((((.((.((.(.((((((-(.(.(.((..((((.......))))..)).)).)))))))).))...)).)))))....((((((....))))))(((((.......))))). ( -39.70, z-score = -1.52, R) >droGri2.scaffold_14624 749641 115 + 4233967 -----UUGCUUAUGUAGCACAUUUG-AACACACUCGUCCAACAAGUCAGGUUGGCCGAGCGGUCUAAGGCGCCAGAUUUAAGCUCUGGUUCUCGAGAGAGAGCGUGGGUUCGAACCCCACA -----..(((((.((.((.(.((((-.((.(.((((.(((((.......))))).)))).))).))))).))...)).)))))....((((((....))))))(((((.......))))). ( -40.10, z-score = -2.00, R) >anoGam1.chr3L 7625644 114 - 41284009 -----UUGUUUGUAUAGCGCUUUUGACUCACAGCCAGC--ACAAGUCAGGAUGGCCGAGCGGUCUAAGGCGCCAGAUUUAAGCUCUGGUUCCCGUAAGGGAGCGUGGGUUCGAACCCCACU -----...........(((((((.(((((.(.((((..--.(......)..)))).).).)))).)))))))((((.......))))((((((....))))))(((((.......))))). ( -42.90, z-score = -1.99, R) >consensus _____UUGCUUAUAUAGCUCUUUUG_A_CAACGACGCCCAACAAGUCAGGUUGGCCGAGCGGUCUAAGGCGCCAGAUUUAAGCUCUGGUUCUCGAGAGAGAGCGUGGGUUCGAACCCCACA ................((..........................(((.....((((....))))...)))((((((.......))))))((((....))))))(((((.......))))). (-29.70 = -29.33 + -0.37)
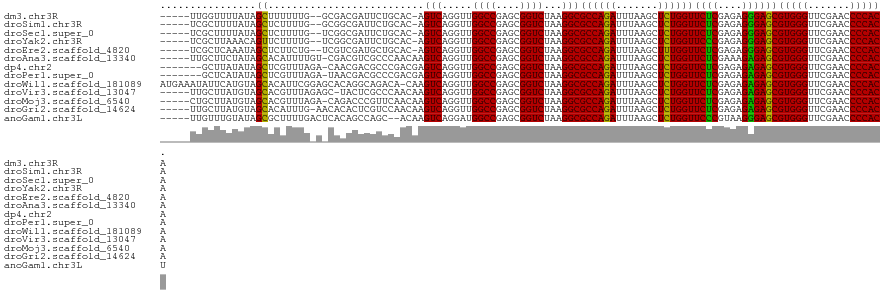
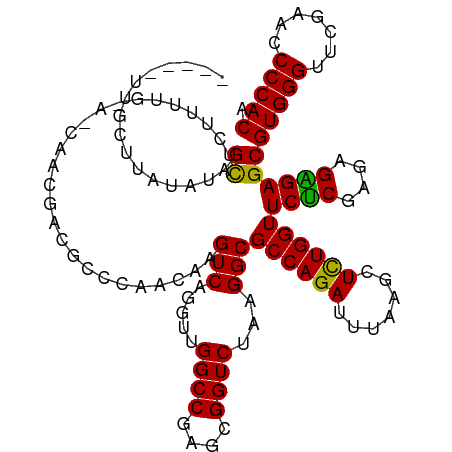
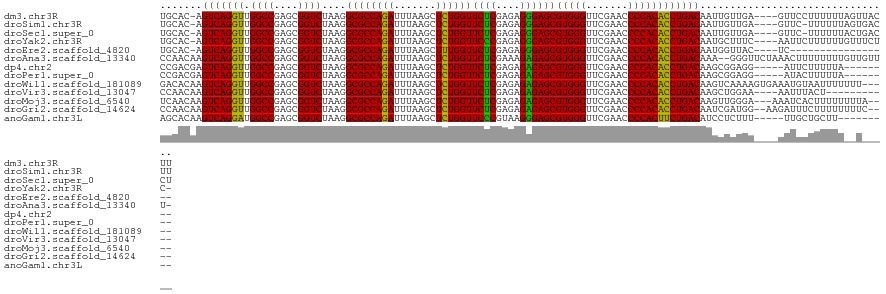
| Location | 20,090,974 – 20,091,091 |
|---|---|
| Length | 117 |
| Sequences | 13 |
| Columns | 122 |
| Reading direction | forward |
| Mean pairwise identity | 81.53 |
| Shannon entropy | 0.41864 |
| G+C content | 0.52453 |
| Mean single sequence MFE | -41.88 |
| Consensus MFE | -39.31 |
| Energy contribution | -38.80 |
| Covariance contribution | -0.51 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.77 |
| SVM RNA-class probability | 0.995143 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
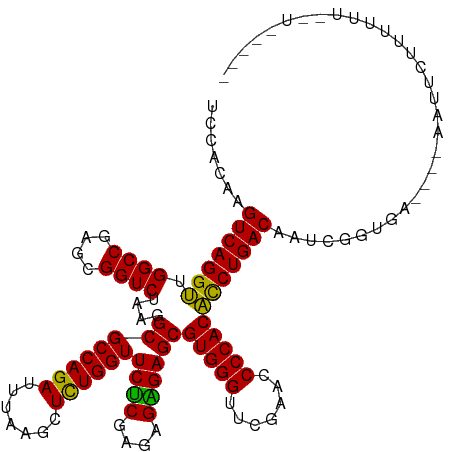
>dm3.chr3R 20090974 117 + 27905053 UGCAC-AGUCAGGUUGGCCGAGCGGUCUAAGGCGCCAGAUUUAAGCUCUGGUUCUCGAGAGGGAGCGUGGGUUCGAACCCCACACCUGACAAUUGUUGA----GUUCCUUUUUUAGUUACUU .....-.(((((((.((((....))))....((((((((.......))))))((((....))))))(((((.......)))))))))))).......((----((..((.....))..)))) ( -40.80, z-score = -1.57, R) >droSim1.chr3R 19923667 116 + 27517382 UGCAC-AGUCAGGUUGGCCGAGCGGUCUAAGGCGCCAGAUUUAAGCUCUGGUUCUCGAGAGGGAGCGUGGGUUCGAACCCCACACCUGACAAUUGUUGA----GUUC-UUUUUUAGUGACUU ..(((-.(((((((.((((....))))....((((((((.......))))))((((....))))))(((((.......))))))))))))......(((----(...-...))))))).... ( -40.20, z-score = -1.26, R) >droSec1.super_0 20423418 116 + 21120651 UGCAC-AGUCAGGUUGGCCGAGCGGUCUAAGGCGCCAGAUUUAAGCUCUGGUUCUCGAGAGGGAGCGUGGGUUCGAACCCCACACCUGACAAUUGUUGA----GUUC-UUUUUUACUGACCU .....-.(((((((.((((....))))....((((((((.......))))))((((....))))))(((((.......))))))))))))....(((.(----((..-......)))))).. ( -39.20, z-score = -0.83, R) >droYak2.chr3R 2416206 116 - 28832112 UGCAC-AGUCAGGUUGGCCGAGCGGUCUAAGGCGCCAGAUUUAAGCUCUGGUUCCCGAGAGGGAGCGUGGGUUCGAACCCCACACCUGACAAUGCUUUC----AAUUCUUUUUUGUUUCUC- .(((.-.(((((((.((((....))))....((((((((.......))))))((((....))))))(((((.......))))))))))))..)))....----..................- ( -44.20, z-score = -2.61, R) >droEre2.scaffold_4820 2498314 100 - 10470090 UGCAC-AGUCAGGUUGGCCGAGCGGUCUAAGGCGCCAGAUUUAAGCUUUGGUUCUCGAGAGGGAGCGUGGGUUCGAACCCCACACCUGACAAUGGUUAC----UC----------------- .....-.(((((((.((((....))))((((((...........))))))((((((....))))))(((((.......)))))))))))).........----..----------------- ( -37.00, z-score = -1.54, R) >droAna3.scaffold_13340 1291746 119 - 23697760 CCAACAAGUCAGGUUGGCCGAGCGGUCUAAGGCGCCAGAUUUAAGCUCUGGUUCUCGAAAGAGAGCGUGGGUUCGAACCCCACACCUGACAAA--GGGUUCUAAACUUUUUUUUGUUGUUU- .(((((((((((((.((((....))))....((((((((.......))))))((((....))))))(((((.......))))))))))))(((--((((.....))))))).))))))...- ( -46.90, z-score = -3.77, R) >dp4.chr2 14106035 109 + 30794189 CCGACGAGUCAGGUUGGCCGAGCGGUCUAAGGCGCCAGAUUUAAGCUCUGGUUCUCGAGAGAGAGCGUGGGUUCGAACCCCACACCUGACAAGCGGAGG-----AUUCUUUUUA-------- ((..((.(((((((.((((....))))....((((((((.......))))))((((....))))))(((((.......))))))))))))...))..))-----..........-------- ( -44.10, z-score = -2.39, R) >droPer1.super_0 5486840 109 - 11822988 CCGACGAGUCAGGUUGGCCGAGCGGUCUAAGGCGCCAGAUUUAAGCUCUGGUUCUCGAGAGAGAGCGUGGGUUCGAACCCCACACCUGACAAGCGGAGG-----AUACUUUUUA-------- ((..((.(((((((.((((....))))....((((((((.......))))))((((....))))))(((((.......))))))))))))...))..))-----..........-------- ( -44.10, z-score = -2.63, R) >droWil1.scaffold_181089 3151141 117 + 12369635 GACACAAGUCAGGUUGGCCGAGCGGUCUAAGGCGCCAGAUUUAAGCUCUGGUUCUCGAGAGAGAGCGUGGGUUCGAACCCCACACCUGACAAGUCAAAAGUGAAAUGUAAUUUUUUU----- (((....(((((((.((((....))))....((((((((.......))))))((((....))))))(((((.......))))))))))))..)))((((((........))))))..----- ( -40.90, z-score = -2.82, R) >droVir3.scaffold_13047 10926534 107 + 19223366 CCAACAAGUCAGGUUGGCCGAGCGGUCUAAGGCGCCAGAUUUAAGCUCUGGUUCUCGAGAGAGAGCGUGGGUUCGAACCCCACACCUGACAAGCUGGAA----AAUUUACU----------- (((.(..(((((((.((((....))))....((((((((.......))))))((((....))))))(((((.......))))))))))))..).)))..----........----------- ( -41.30, z-score = -2.84, R) >droMoj3.scaffold_6540 3299160 115 - 34148556 UCAACAAGUCAGGUUGGCCGAGCGGUCUAAGGCGCCAGAUUUAAGCUCUGGUUCUCGAGAGAGAGCGUGGGUUCGAACCCCACACCUGACAAGUUGGGA---AAAUCACUUUUUUUUA---- .((((..(((((((.((((....))))....((((((((.......))))))((((....))))))(((((.......))))))))))))..))))(((---(((......)))))).---- ( -43.30, z-score = -3.01, R) >droGri2.scaffold_14624 749672 116 + 4233967 CCAACAAGUCAGGUUGGCCGAGCGGUCUAAGGCGCCAGAUUUAAGCUCUGGUUCUCGAGAGAGAGCGUGGGUUCGAACCCCACACCUGACAAUCGAUGG--AAGAUUUCUUUUUUUUC---- .......(((((((.((((....))))....((((((((.......))))))((((....))))))(((((.......))))))))))))....((..(--(((....))))..))..---- ( -40.40, z-score = -1.97, R) >anoGam1.chr3L 7625674 108 - 41284009 AGCACAAGUCAGGAUGGCCGAGCGGUCUAAGGCGCCAGAUUUAAGCUCUGGUUCCCGUAAGGGAGCGUGGGUUCGAACCCCACUUCUGACAUCCUCUUU-----UUGCUGCUU--------- ((((((((..(((((((((...........)))..((((...........((((((....))))))(((((.......))))).)))).))))))...)-----))).)))).--------- ( -42.00, z-score = -2.09, R) >consensus UCCACAAGUCAGGUUGGCCGAGCGGUCUAAGGCGCCAGAUUUAAGCUCUGGUUCUCGAGAGAGAGCGUGGGUUCGAACCCCACACCUGACAAUCGGUGA____AAUUCUUUUUU__U_____ .......(((((((.((((....))))....((((((((.......))))))((((....))))))(((((.......))))))))))))................................ (-39.31 = -38.80 + -0.51)
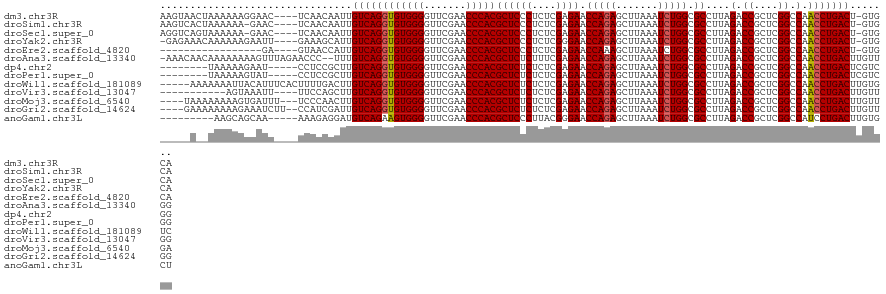
| Location | 20,090,974 – 20,091,091 |
|---|---|
| Length | 117 |
| Sequences | 13 |
| Columns | 122 |
| Reading direction | reverse |
| Mean pairwise identity | 81.53 |
| Shannon entropy | 0.41864 |
| G+C content | 0.52453 |
| Mean single sequence MFE | -34.93 |
| Consensus MFE | -29.84 |
| Energy contribution | -29.99 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.948377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 20090974 117 - 27905053 AAGUAACUAAAAAAGGAAC----UCAACAAUUGUCAGGUGUGGGGUUCGAACCCACGCUCCCUCUCGAGAACCAGAGCUUAAAUCUGGCGCCUUAGACCGCUCGGCCAACCUGACU-GUGCA .(((..((.....))..))----)...((...((((((((((((.......)))))........(((((..(((((.......)))))(......)....)))))...))))))).-.)).. ( -31.50, z-score = -0.35, R) >droSim1.chr3R 19923667 116 - 27517382 AAGUCACUAAAAAA-GAAC----UCAACAAUUGUCAGGUGUGGGGUUCGAACCCACGCUCCCUCUCGAGAACCAGAGCUUAAAUCUGGCGCCUUAGACCGCUCGGCCAACCUGACU-GUGCA ..(.(((.......-(...----.).......((((((((((((.......)))))........(((((..(((((.......)))))(......)....)))))...))))))).-)))). ( -30.60, z-score = -0.32, R) >droSec1.super_0 20423418 116 - 21120651 AGGUCAGUAAAAAA-GAAC----UCAACAAUUGUCAGGUGUGGGGUUCGAACCCACGCUCCCUCUCGAGAACCAGAGCUUAAAUCUGGCGCCUUAGACCGCUCGGCCAACCUGACU-GUGCA .(((((((......-..))----)........((((((((((((.......)))))((((..((....))....))))....)))))))......))))((.(((.(.....).))-).)). ( -33.20, z-score = -0.33, R) >droYak2.chr3R 2416206 116 + 28832112 -GAGAAACAAAAAAGAAUU----GAAAGCAUUGUCAGGUGUGGGGUUCGAACCCACGCUCCCUCUCGGGAACCAGAGCUUAAAUCUGGCGCCUUAGACCGCUCGGCCAACCUGACU-GUGCA -..................----....((((.((((((((((((.......)))))(((...(((.(((..(((((.......)))))..))).)))......)))..))))))).-)))). ( -41.70, z-score = -2.62, R) >droEre2.scaffold_4820 2498314 100 + 10470090 -----------------GA----GUAACCAUUGUCAGGUGUGGGGUUCGAACCCACGCUCCCUCUCGAGAACCAAAGCUUAAAUCUGGCGCCUUAGACCGCUCGGCCAACCUGACU-GUGCA -----------------..----.....(((.((((((((((((.......)))))........(((((.......(((.......)))...........)))))...))))))).-))).. ( -28.37, z-score = -0.17, R) >droAna3.scaffold_13340 1291746 119 + 23697760 -AAACAACAAAAAAAAGUUUAGAACCC--UUUGUCAGGUGUGGGGUUCGAACCCACGCUCUCUUUCGAGAACCAGAGCUUAAAUCUGGCGCCUUAGACCGCUCGGCCAACCUGACUUGUUGG -...((((((...((((.........)--)))((((((((((((.......)))))((((((....)))).(((((.......))))).))....(.((....)).).))))))))))))). ( -36.60, z-score = -1.95, R) >dp4.chr2 14106035 109 - 30794189 --------UAAAAAGAAU-----CCUCCGCUUGUCAGGUGUGGGGUUCGAACCCACGCUCUCUCUCGAGAACCAGAGCUUAAAUCUGGCGCCUUAGACCGCUCGGCCAACCUGACUCGUCGG --------..........-----...((((..((((((((((((.......)))))((((((....)))).(((((.......))))).))....(.((....)).).)))))))..).))) ( -35.50, z-score = -1.51, R) >droPer1.super_0 5486840 109 + 11822988 --------UAAAAAGUAU-----CCUCCGCUUGUCAGGUGUGGGGUUCGAACCCACGCUCUCUCUCGAGAACCAGAGCUUAAAUCUGGCGCCUUAGACCGCUCGGCCAACCUGACUCGUCGG --------..........-----...((((..((((((((((((.......)))))((((((....)))).(((((.......))))).))....(.((....)).).)))))))..).))) ( -35.50, z-score = -1.53, R) >droWil1.scaffold_181089 3151141 117 - 12369635 -----AAAAAAAUUACAUUUCACUUUUGACUUGUCAGGUGUGGGGUUCGAACCCACGCUCUCUCUCGAGAACCAGAGCUUAAAUCUGGCGCCUUAGACCGCUCGGCCAACCUGACUUGUGUC -----.......................((..((((((((((((.......)))))((((((....)))).(((((.......))))).))....(.((....)).).)))))))..))... ( -33.80, z-score = -1.77, R) >droVir3.scaffold_13047 10926534 107 - 19223366 -----------AGUAAAUU----UUCCAGCUUGUCAGGUGUGGGGUUCGAACCCACGCUCUCUCUCGAGAACCAGAGCUUAAAUCUGGCGCCUUAGACCGCUCGGCCAACCUGACUUGUUGG -----------........----..(((((..((((((((((((.......)))))((((((....)))).(((((.......))))).))....(.((....)).).)))))))..))))) ( -39.60, z-score = -2.77, R) >droMoj3.scaffold_6540 3299160 115 + 34148556 ----UAAAAAAAAGUGAUUU---UCCCAACUUGUCAGGUGUGGGGUUCGAACCCACGCUCUCUCUCGAGAACCAGAGCUUAAAUCUGGCGCCUUAGACCGCUCGGCCAACCUGACUUGUUGA ----................---...((((..((((((((((((.......)))))((((((....)))).(((((.......))))).))....(.((....)).).)))))))..)))). ( -37.60, z-score = -2.53, R) >droGri2.scaffold_14624 749672 116 - 4233967 ----GAAAAAAAAGAAAUCUU--CCAUCGAUUGUCAGGUGUGGGGUUCGAACCCACGCUCUCUCUCGAGAACCAGAGCUUAAAUCUGGCGCCUUAGACCGCUCGGCCAACCUGACUUGUUGG ----.................--(((.(((..((((((((((((.......)))))((((((....)))).(((((.......))))).))....(.((....)).).)))))))))).))) ( -34.50, z-score = -1.36, R) >anoGam1.chr3L 7625674 108 + 41284009 ---------AAGCAGCAA-----AAAGAGGAUGUCAGAAGUGGGGUUCGAACCCACGCUCCCUUACGGGAACCAGAGCUUAAAUCUGGCGCCUUAGACCGCUCGGCCAUCCUGACUUGUGCU ---------.((((.(((-----....((((((((....(((((.......)))))((((((....)))).(((((.......)))))...........))..)).))))))...))))))) ( -35.60, z-score = -0.87, R) >consensus _____A__AAAAAAGAAUU____UCAACAAUUGUCAGGUGUGGGGUUCGAACCCACGCUCUCUCUCGAGAACCAGAGCUUAAAUCUGGCGCCUUAGACCGCUCGGCCAACCUGACUUGUGCA ................................((((((((((((.......)))))((((((....)))).(((((.......))))).))....(.((....)).).)))))))....... (-29.84 = -29.99 + 0.15)
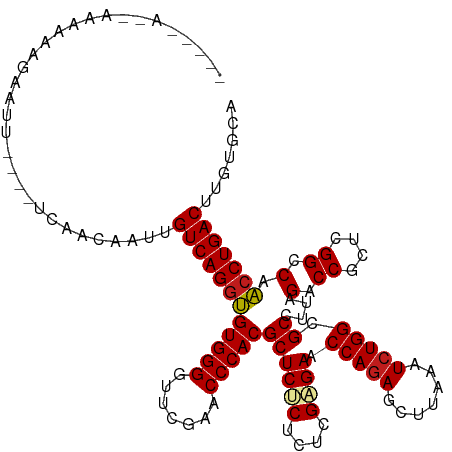
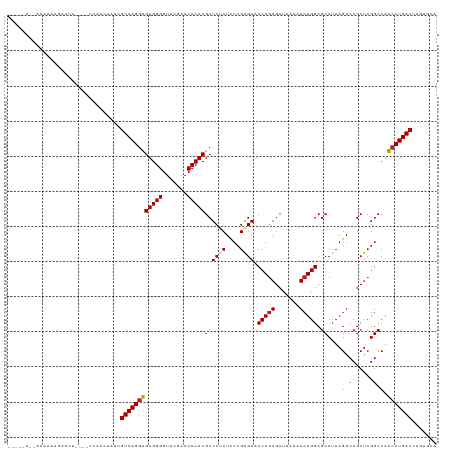
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:40:11 2011