
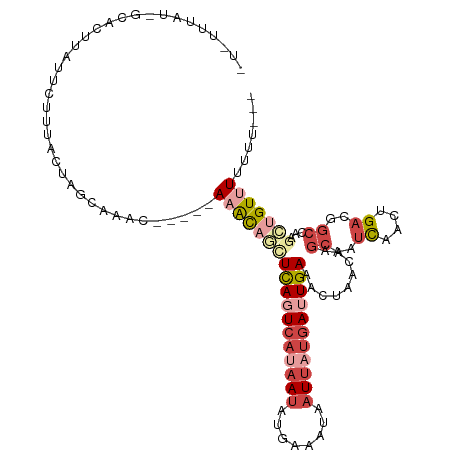
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 20,010,033 – 20,010,160 |
| Length | 127 |
| Max. P | 0.977484 |

| Location | 20,010,033 – 20,010,142 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 67.97 |
| Shannon entropy | 0.59159 |
| G+C content | 0.31605 |
| Mean single sequence MFE | -17.76 |
| Consensus MFE | -8.97 |
| Energy contribution | -11.22 |
| Covariance contribution | 2.25 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.977484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 20010033 109 + 27905053 AUGUGCAU-AAGCU-AUUCUUUACUAGGAAAC-----AAGCAGUUCAGUCACAAUAUGAAAUAAUUAUGAUUGAAACUAACAAGCAAUCAACUGACGGCCAAGCUGUUUUUUUUUU ...(((..-..(((-.(((((....)))))..-----.)))((((((((((.(((........))).)))))).)))).....))).......((((((...))))))........ ( -20.40, z-score = -0.68, R) >droSim1.chr3R 19843462 106 + 27517382 -UAUUUAU-GCACUUAUUCUUUACUAGCAAAC-----AAACAGCUCAGUCAUAAUAUGAAAUAAUUAUGAUUGAAACUAACAAGCAAUCAACUGACGGCCAAGCUGUUUUUUU--- -......(-((...............)))...-----((((((((((((((((((........))))))))))).........((..((....))..))...)))))))....--- ( -21.86, z-score = -2.86, R) >droSec1.super_0 20347926 91 + 21120651 -----------------UCUUUACUAGCAAAC-----AAACAGCUCAGUCAUAAUAUGAAAUAAUUAUGAUUGAAACUAACAAGCAAUCAACUGACGGCCAAGCUGUUUUUUU--- -----------------...............-----((((((((((((((((((........))))))))))).........((..((....))..))...)))))))....--- ( -21.20, z-score = -3.68, R) >droYak2.chr3R 2339399 104 - 28832112 -UAUUUAU-GCACCUCAACCUCACUAAAAACC-----AAGCAUAUCAUUCAUAAUAUUAAAUAAUUAUGAUUGAAACUAACAAGCAAUCAACUGACGGCCAAGCUGUGUUU----- -.......-..................((((.-----.(((...(((.(((((((........))))))).))).........((..((....))..))...)))..))))----- ( -14.50, z-score = -1.39, R) >droEre2.scaffold_4820 2422542 94 - 10470090 ----------UAUUUAUCCUACACUAACCACC-----AAGC-UCUCAGUCAUAAUAUUAAAUAAUUAUGAUUGAAACUAACAAGCAAUCAACUGACGGCCAU-CUGUUUUU----- ----------......................-----((((-..(((((((((((........))))))))))).........((..((....))..))...-..))))..----- ( -13.40, z-score = -2.25, R) >droAna3.scaffold_13340 1213108 110 - 23697760 ACCUUUGUAGUACAAAGUUCGUAUUACAAAUUGCCAAAAAUUACUUAGUACAAACACUAAAAAAUCAUAAAUGAAAAUUCCCAAGAGUUAGC-GCCGAC--GGUUGUGUUUUU--- ...((((((((((.......))))))))))..(((.........(((((......)))))............(..(((((....)))))..)-......--))).........--- ( -15.20, z-score = 0.02, R) >consensus _U_UUUAU_GCACUUAUUCUUUACUAGCAAAC_____AAACAGCUCAGUCAUAAUAUGAAAUAAUUAUGAUUGAAACUAACAAGCAAUCAACUGACGGCCAAGCUGUUUUUUU___ .....................................((((((((((((((((((........))))))))))).........((..((....))..))...)))))))....... ( -8.97 = -11.22 + 2.25)

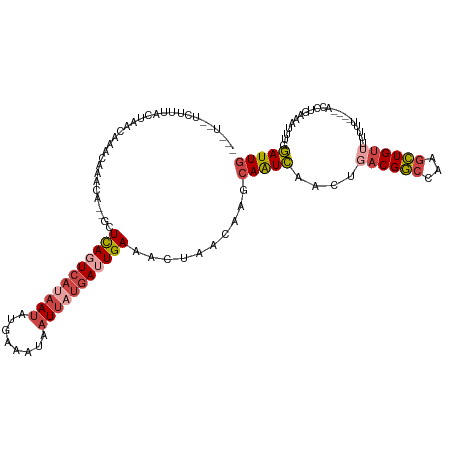
| Location | 20,010,042 – 20,010,160 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.87 |
| Shannon entropy | 0.49567 |
| G+C content | 0.32158 |
| Mean single sequence MFE | -20.27 |
| Consensus MFE | -9.62 |
| Energy contribution | -10.96 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.798844 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 20010042 118 + 27905053 AGCUAUUCUUUACUAGGAAACAAGCA--GUUCAGUCACAAUAUGAAAUAAUUAUGAUUGAAACUAACAAGCAAUCAACUGACGGCCAAGCUGUUUUUUUUUUUACCUGAAAUUUGGAUUG .(((.(((((....)))))...)))(--(((((((((.(((........))).)))))).))))......(((((....((((((...))))))...((((......))))....))))) ( -19.70, z-score = -0.16, R) >droSim1.chr3R 19843473 113 + 27517382 --UUAUUCUUUACUAGCAAACAAACA--GCUCAGUCAUAAUAUGAAAUAAUUAUGAUUGAAACUAACAAGCAAUCAACUGACGGCCAAGCUGUUUUUUUA---ACCUGAAAUUUGGAUUG --...............(((.(((((--(((((((((((((........))))))))))).........((..((....))..))...))))))).))).---.((........)).... ( -23.20, z-score = -2.71, R) >droSec1.super_0 20347926 109 + 21120651 ------UCUUUACUAGCAAACAAACA--GCUCAGUCAUAAUAUGAAAUAAUUAUGAUUGAAACUAACAAGCAAUCAACUGACGGCCAAGCUGUUUUUUUA---AACUGAAAUUUGGAUUG ------...........(((.(((((--(((((((((((((........))))))))))).........((..((....))..))...))))))).))).---................. ( -21.90, z-score = -2.44, R) >droYak2.chr3R 2339410 111 - 28832112 --CUCAACCUCACUAAAAACCAAGCA--UAUCAUUCAUAAUAUUAAAUAAUUAUGAUUGAAACUAACAAGCAAUCAACUGACGGCCAAGCUGUGUUUU-----AGCUGAAAUUUGGAUUG --.((((..(((((((((....(((.--..(((.(((((((........))))))).))).........((..((....))..))...)))...))))-----)).)))...)))).... ( -21.00, z-score = -2.40, R) >droEre2.scaffold_4820 2422549 105 - 10470090 ------CCUACACUAACCACCAAGC---UCUCAGUCAUAAUAUUAAAUAAUUAUGAUUGAAACUAACAAGCAAUCAACUGACGGCCAU-CUGUUUUUG-----AGCUAAAAUUUGGAUUG ------.............(((((.---..(((((((((((........)))))))))))........(((..((((..(((((....-))))).)))-----))))....))))).... ( -22.10, z-score = -3.47, R) >droAna3.scaffold_13340 1213122 113 - 23697760 AAGUUCGUAUUACAAAUUGCCAAAAAUUACUUAGUACAAACACUAAAAAAUCAUAAAUGAAAAUUCCCAAGAGUUAGC-GCCGAC--GGUUGUGUUUUU----GUUCCAAAUUCGAAUUG .(((((((((((..((((......))))...))))))((((((...............(..(((((....)))))..)-(((...--))).))))))..----...........))))). ( -13.70, z-score = 0.54, R) >consensus ___U__UCUUUACUAACAAACAAACA__GCUCAGUCAUAAUAUGAAAUAAUUAUGAUUGAAACUAACAAGCAAUCAACUGACGGCCAAGCUGUUUUUUU____ACCUGAAAUUUGGAUUG ..............................(((((((((((........)))))))))))..........(((((....((((((...)))))).....................))))) ( -9.62 = -10.96 + 1.34)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:39:58 2011