
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,481,770 – 8,481,862 |
| Length | 92 |
| Max. P | 0.991777 |

| Location | 8,481,770 – 8,481,862 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 72.66 |
| Shannon entropy | 0.62702 |
| G+C content | 0.53284 |
| Mean single sequence MFE | -29.82 |
| Consensus MFE | -25.93 |
| Energy contribution | -25.23 |
| Covariance contribution | -0.70 |
| Combinations/Pair | 1.62 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.991777 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
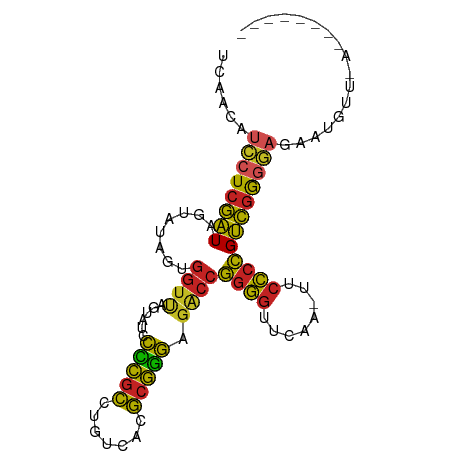
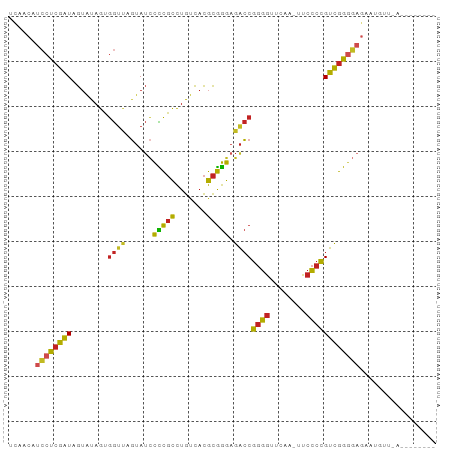
>dm3.chr2L 8481770 92 + 23011544 UCAACGUCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAA-UUCCCCGUCGGGGAGACGAUUGUCAAUAC-- ......((((((((........((((......(((((.......))))).))))((((.....-..))))))))))))(((....))).....-- ( -31.90, z-score = -0.94, R) >droSim1.chr2L 13569553 80 - 22036055 UCAACAUCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAA-UUCCCCGUCGGGGAGAA-------------- ......((((((((........((((......(((((.......))))).))))((((.....-..))))))))))))...-------------- ( -30.00, z-score = -1.49, R) >droSec1.super_3 3055833 88 - 7220098 UCAACAUCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAA-UUCCCCGUCGGGGAGAAUGUGAAAG------ (((...((((((((........((((......(((((.......))))).))))((((.....-..)))))))))))).....)))...------ ( -31.70, z-score = -1.77, R) >droYak2.chr2L 11127884 93 + 22324452 CUAACGUCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAA-UUCCCCGUCGGGGAGACAAUUGUGCGAACU- ...............(((((((.(((....(((((.((((.....)))).(((.((((.....-..))))))))))))))).))))))).....- ( -32.60, z-score = -0.91, R) >droEre2.scaffold_4929 9074763 89 + 26641161 UCAACGUCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAA-UUCCCCGUCGGGGAGAAUGUUAAAAC----- ..(((((.(((((((.((.......)).))))(((.((((.....)))).(((.((((.....-..))))))))))))).))))).....----- ( -30.60, z-score = -1.34, R) >droPer1.super_1 1798461 89 - 10282868 UCAACAUCCUCGAUAGUAUAGUGGUCAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAA-UUCCCCGUCGGGGAGAGAUGUAAAAC----- ...(((((.(((((.........)))....(((((.((((.....)))).(((.((((.....-..))))))))))))))))))).....----- ( -31.90, z-score = -1.71, R) >droWil1.scaffold_180772 810413 84 - 8906247 UAACGGAACUAAAUUGAUUAGAAAGUAUUAAUUUUUUUUGGUCUAUAAAAGAACGUAAAGCAAAUUAUCCGUUAAAUGAGAUAC----------- (((((((.((((.....))))...((.(((..((((((((.....))))))))..))).))......)))))))..........----------- ( -13.90, z-score = -1.70, R) >droMoj3.scaffold_6500 7169180 94 - 32352404 ACAACAUCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAA-UUCCCCGUCGGGGAGAUGUGUGAUUUUUUUU (((...((((((((........((((......(((((.......))))).))))((((.....-..))))))))))))....))).......... ( -32.00, z-score = -1.46, R) >droVir3.scaffold_12963 5696911 88 + 20206255 UCAACAUCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAA-UUCCCCGUCGGGGAGAAUGUUCAAA------ ..(((((.(((((((.((.......)).))))(((.((((.....)))).(((.((((.....-..))))))))))))).)))))....------ ( -30.40, z-score = -1.47, R) >droGri2.scaffold_15252 9009322 72 - 17193109 ---------UCGGUGGUUCAGUGGU-AGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGA-UUCCCGGCCGAUGCAUAUC------------ ---------..((((.....(((((-((.........)))))))(((..(((((.(((.....-..)))))))).))).))))------------ ( -29.20, z-score = -0.40, R) >anoGam1.chr3L 4289357 83 + 41284009 -CAACGUCCUCGAUAGUAUAGUGGUCAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCGA-UUCCCCGUCGGGGAGGAAGAA---------- -...(.((((((((........((((......(((((.......))))).))))((((.....-..)))))))))))).).....---------- ( -33.80, z-score = -1.42, R) >consensus UCAACAUCCUCGAUAGUAUAGUGGUUAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAA_UUCCCCGUCGGGGAGAAUGUU_A________ ......((((((((........((((......(((((.......))))).))))((((........))))))))))))................. (-25.93 = -25.23 + -0.70)

| Location | 8,481,770 – 8,481,862 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 72.66 |
| Shannon entropy | 0.62702 |
| G+C content | 0.53284 |
| Mean single sequence MFE | -27.16 |
| Consensus MFE | -22.64 |
| Energy contribution | -21.70 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.71 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.982476 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
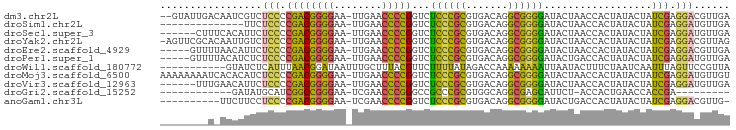
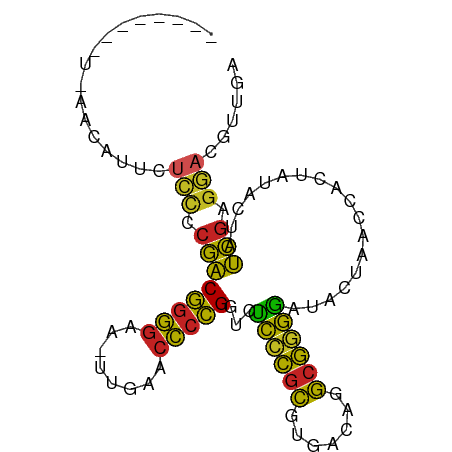
>dm3.chr2L 8481770 92 - 23011544 --GUAUUGACAAUCGUCUCCCCGACGGGGAA-UUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGACGUUGA --.......(((.(((((...((((((((..-.....)))))(((.(((((.......))))))))................))).)))))))). ( -30.50, z-score = -0.71, R) >droSim1.chr2L 13569553 80 + 22036055 --------------UUCUCCCCGACGGGGAA-UUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGAUGUUGA --------------...(((.((((((((..-.....)))))(((.(((((.......))))))))................))).)))...... ( -27.40, z-score = -0.84, R) >droSec1.super_3 3055833 88 + 7220098 ------CUUUCACAUUCUCCCCGACGGGGAA-UUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGAUGUUGA ------...((((((((((.....(((((..-.....)))))(((.(((((.......))))))))..................))))))).))) ( -30.90, z-score = -1.85, R) >droYak2.chr2L 11127884 93 - 22324452 -AGUUCGCACAAUUGUCUCCCCGACGGGGAA-UUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGACGUUAG -................(((.((((((((..-.....)))))(((.(((((.......))))))))................))).)))...... ( -27.30, z-score = 0.13, R) >droEre2.scaffold_4929 9074763 89 - 26641161 -----GUUUUAACAUUCUCCCCGACGGGGAA-UUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGACGUUGA -----...(((((.(((((.....(((((..-.....)))))(((.(((((.......))))))))..................))))).))))) ( -29.10, z-score = -1.13, R) >droPer1.super_1 1798461 89 + 10282868 -----GUUUUACAUCUCUCCCCGACGGGGAA-UUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUGACCACUAUACUAUCGAGGAUGUUGA -----.....(((((.(((......((((..-.....))))((((((((((.......)))))).....))))...........))))))))... ( -30.20, z-score = -1.16, R) >droWil1.scaffold_180772 810413 84 + 8906247 -----------GUAUCUCAUUUAACGGAUAAUUUGCUUUACGUUCUUUUAUAGACCAAAAAAAAUUAAUACUUUCUAAUCAAUUUAGUUCCGUUA -----------..........(((((((((((((..(((..(.(((.....))).))))..)))))).......((((.....)))).))))))) ( -9.60, z-score = -1.29, R) >droMoj3.scaffold_6500 7169180 94 + 32352404 AAAAAAAAUCACACAUCUCCCCGACGGGGAA-UUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGAUGUUGU ............(((.((((.((((((((..-.....)))))(((.(((((.......))))))))................))).))).).))) ( -28.60, z-score = -1.23, R) >droVir3.scaffold_12963 5696911 88 - 20206255 ------UUUGAACAUUCUCCCCGACGGGGAA-UUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGAUGUUGA ------....(((((((((.....(((((..-.....)))))(((.(((((.......))))))))..................))))))))).. ( -32.00, z-score = -2.25, R) >droGri2.scaffold_15252 9009322 72 + 17193109 ------------GAUAUGCAUCGGCCGGGAA-UCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCU-ACCACUGAACCACCGA--------- ------------........(((((((((..-.....)))))((((((.((...)).)))).)).....-............))))--------- ( -24.20, z-score = -0.01, R) >anoGam1.chr3L 4289357 83 - 41284009 ----------UUCUUCCUCCCCGACGGGGAA-UCGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUGACCACUAUACUAUCGAGGACGUUG- ----------....(((((......((((..-.....))))((((((((((.......)))))).....))))...........))))).....- ( -29.00, z-score = -0.65, R) >consensus ________U_AACAUUCUCCCCGACGGGGAA_UUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUAACCACUAUACUAUCGAGGACGUUGA .................(((.((((((((........)))))...((((((.......))))))..................))).)))...... (-22.64 = -21.70 + -0.94)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:26:12 2011