| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,953,673 – 19,953,767 |
| Length | 94 |
| Max. P | 0.985898 |

| Location | 19,953,673 – 19,953,767 |
|---|---|
| Length | 94 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 70.04 |
| Shannon entropy | 0.56561 |
| G+C content | 0.40163 |
| Mean single sequence MFE | -19.12 |
| Consensus MFE | -12.56 |
| Energy contribution | -12.00 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.985898 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
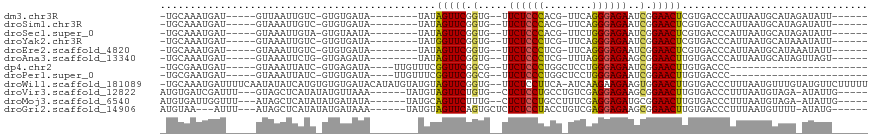
>dm3.chr3R 19953673 94 + 27905053 ------AAUAUCUAUGCAUUAAUGGGUCACGAGUUCCGAUUCUCCCUGAA-CGUGGGAGAA--CACCGAACUAUA--------UAUCACAC-GACAAUUAAC-----AUCAUUUGCA- ------........((((..(((((......(((((.(.(((((((....-...)))))))--..).)))))...--------..((....-))........-----.)))))))))- ( -19.20, z-score = -0.96, R) >droSim1.chr3R 19784948 94 + 27517382 ------AAUAUCUAUGCAUUAAUGGGUCACGAGUUCCGAUUCUCCCUGAA-CGUGGGAGAA--CACCGAACUAUA--------UAUCACAC-GACAAUUUAC-----AUCAUUUGCA- ------........((((..(((((......(((((.(.(((((((....-...)))))))--..).)))))...--------..((....-))........-----.)))))))))- ( -19.20, z-score = -0.95, R) >droSec1.super_0 20291715 94 + 21120651 ------AAUAUCUAUGCAUUAAUGGGUCACGAGUUCCGAUUCUCCCAGAA-CGUGGGAGAA--CACCGAACUAUA--------UAUUACAC-UACAAUUUAC-----AUCAUUUGCA- ------........((((..(((((....).(((((.(.((((((((...-..))))))))--..).)))))...--------........-..........-----..))))))))- ( -19.00, z-score = -1.53, R) >droYak2.chr3R 2282649 94 - 28832112 ------AAUAUUUAUGCAUUAAUGGGUCACGAGUUCCGAUUCUCCCUGAA-CGAGGGAGAA--CACCGAACCAUA--------UAUCACAC-GACAAUUUAC-----AUCAUUUGCA- ------........((((...((((.((.((.....)).((((((((...-..))))))))--....)).)))).--------..((....-))........-----......))))- ( -19.20, z-score = -1.30, R) >droEre2.scaffold_4820 2364629 94 - 10470090 ------AAUAUUUAUGCAUUAAUGGGUCACGAGUUCCGAUUCUCCCUGAA-CGAGGGAGAA--CACCGAACUAUA--------UAUCACAC-GACAAUUUAC-----AUCAUUUGCA- ------........((((..(((((......(((((.(.((((((((...-..))))))))--..).)))))...--------..((....-))........-----.)))))))))- ( -20.30, z-score = -1.83, R) >droAna3.scaffold_13340 1154998 94 - 23697760 ------ACUAACUAUGCAUUAAUGGGUCACAAGUUCCGCUUCUCCCUAAA-CGAGGGAGAA--CACCGAACUAUA--------UAUCUCAC-CAGAAUUUAC-----AUCAUUUGCA- ------........((((..(((((......(((((.(.((((((((...-..))))))))--..).)))))...--------..(((...-.)))......-----.)))))))))- ( -21.00, z-score = -2.73, R) >dp4.chr2 14486358 82 + 30794189 -----------------------GGGUCACAAGUUCCGAUUCUCCCAGGAGCCAGGGAGAA--CGCCGAACCGAAACAA----UAUCUCAC-GAUAAUUUAC-----AUCAUUCGCA- -----------------------(..((....((((((.(((((((.(....).)))))))--))..)))).))..)..----((((....-))))......-----..........- ( -17.20, z-score = -0.63, R) >droPer1.super_0 5887416 82 - 11822988 -----------------------GGGUCACAAGUUCCGAUUCUCCCAGGAGCCAGGGAGAA--CGCCGAACCGAAACAA----UAUCACAC-GAUAAUUUAC-----AUCAUUCGCA- -----------------------(..((....((((((.(((((((.(....).)))))))--))..)))).))..)..----((((....-))))......-----..........- ( -17.30, z-score = -1.02, R) >droWil1.scaffold_181089 6414890 114 - 12369635 AAAAAGAACAUACAAACAUUAAAGGGUCACAAGUUCCACUUCUUCUUGAU-UGAAGGAGAA--CACCGAACUACAUACAUAUGUAUCACACACAUGAUAUAUUGAAAAUCAUUUGCA- .......................((.(((..(((((...((((((((...-..))))))))--....)))))........((((((((......)))))))))))...)).......- ( -17.50, z-score = -0.84, R) >droVir3.scaffold_12822 1097576 101 - 4096053 -----CAAUAU-UCUACAUUAAAGGGUCACAAGUUCCGCUUCUCCUCGACAGGCAGGAGAG--CACAGAACUACAUA------UUUAACAUAUAUGAGCUAC---AAAUCGAUCACAU -----......-............((((...(((((.(((.((((((.....).)))))))--)...))))).((((------(.......)))))......---.....)))).... ( -20.50, z-score = -2.79, R) >droMoj3.scaffold_6540 7399916 101 + 34148556 -----CAAUAU-UCUACAUUAAAGGGUCACAAGUUCCGCAUCUCCUCGAAAGGCAGGAGAG--CAAAGAACUGCAUA------UAUAUCAUAUAUGAGCUAU---AAACCAAUCACAU -----......-............(((....(((((.((.(((((((....)..)))))))--)...))))).((((------(((...)))))))......---..)))........ ( -21.30, z-score = -2.59, R) >droGri2.scaffold_14906 9634535 99 - 14172833 ------CAUAU-AAAACAUUAAAGGGUCACAAGUUCCGCUUCUCCUCGACAGGUAGGAGAGAGCACUGAACUACAUA------UUUAUCAUAUAUGAGCUAU---AAAU---UUACAU ------(((((-(......((((..((....(((((.((((((((((.....).)))))).)))...)))))))...------))))...))))))......---....---...... ( -17.70, z-score = -1.22, R) >consensus ______AAUAU_UAUGCAUUAAUGGGUCACAAGUUCCGAUUCUCCCUGAA_CGAGGGAGAA__CACCGAACUAUA________UAUCACAC_GAUAAUUUAC_____AUCAUUUGCA_ ................................((((...(((((((........)))))))......))))............................................... (-12.56 = -12.00 + -0.56)
| Location | 19,953,673 – 19,953,767 |
|---|---|
| Length | 94 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 70.04 |
| Shannon entropy | 0.56561 |
| G+C content | 0.40163 |
| Mean single sequence MFE | -24.58 |
| Consensus MFE | -12.95 |
| Energy contribution | -12.42 |
| Covariance contribution | -0.53 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.967940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
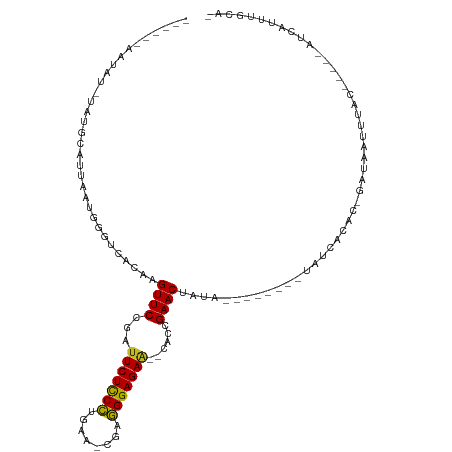
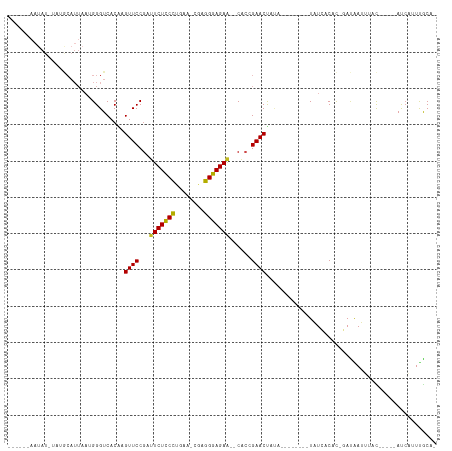
>dm3.chr3R 19953673 94 - 27905053 -UGCAAAUGAU-----GUUAAUUGUC-GUGUGAUA--------UAUAGUUCGGUG--UUCUCCCACG-UUCAGGGAGAAUCGGAACUCGUGACCCAUUAAUGCAUAGAUAUU------ -((((((((..-----((((..((((-....))))--------...(((((.(.(--(((((((...-....))))))))).)))))..)))).))))..))))........------ ( -24.70, z-score = -1.33, R) >droSim1.chr3R 19784948 94 - 27517382 -UGCAAAUGAU-----GUAAAUUGUC-GUGUGAUA--------UAUAGUUCGGUG--UUCUCCCACG-UUCAGGGAGAAUCGGAACUCGUGACCCAUUAAUGCAUAGAUAUU------ -((((..((((-----(.....((((-....))))--------((((((((.(.(--(((((((...-....))))))))).))))).)))...))))).))))........------ ( -24.50, z-score = -1.33, R) >droSec1.super_0 20291715 94 - 21120651 -UGCAAAUGAU-----GUAAAUUGUA-GUGUAAUA--------UAUAGUUCGGUG--UUCUCCCACG-UUCUGGGAGAAUCGGAACUCGUGACCCAUUAAUGCAUAGAUAUU------ -((((..((((-----(.....((((-......))--------)).(((((.(.(--((((((((..-...)))))))))).))))).......))))).))))........------ ( -24.30, z-score = -1.81, R) >droYak2.chr3R 2282649 94 + 28832112 -UGCAAAUGAU-----GUAAAUUGUC-GUGUGAUA--------UAUGGUUCGGUG--UUCUCCCUCG-UUCAGGGAGAAUCGGAACUCGUGACCCAUUAAUGCAUAAAUAUU------ -((((..((((-----(.....((((-....))))--------((((((((.(.(--((((((((..-...)))))))))).)))).))))...))))).))))........------ ( -27.10, z-score = -2.32, R) >droEre2.scaffold_4820 2364629 94 + 10470090 -UGCAAAUGAU-----GUAAAUUGUC-GUGUGAUA--------UAUAGUUCGGUG--UUCUCCCUCG-UUCAGGGAGAAUCGGAACUCGUGACCCAUUAAUGCAUAAAUAUU------ -((((..((((-----(.....((((-....))))--------((((((((.(.(--((((((((..-...)))))))))).))))).)))...))))).))))........------ ( -25.50, z-score = -2.19, R) >droAna3.scaffold_13340 1154998 94 + 23697760 -UGCAAAUGAU-----GUAAAUUCUG-GUGAGAUA--------UAUAGUUCGGUG--UUCUCCCUCG-UUUAGGGAGAAGCGGAACUUGUGACCCAUUAAUGCAUAGUUAGU------ -.((.(((.((-----(((.....((-(((.(.((--------((.(((((.((.--((((((((..-...)))))))))).))))))))).).))))).))))).))).))------ ( -26.30, z-score = -2.21, R) >dp4.chr2 14486358 82 - 30794189 -UGCGAAUGAU-----GUAAAUUAUC-GUGAGAUA----UUGUUUCGGUUCGGCG--UUCUCCCUGGCUCCUGGGAGAAUCGGAACUUGUGACCC----------------------- -.((((.(((.-----.(((..((((-....))))----)))..)))((((.(.(--(((((((.(....).))))))))).)))))))).....----------------------- ( -25.70, z-score = -1.85, R) >droPer1.super_0 5887416 82 + 11822988 -UGCGAAUGAU-----GUAAAUUAUC-GUGUGAUA----UUGUUUCGGUUCGGCG--UUCUCCCUGGCUCCUGGGAGAAUCGGAACUUGUGACCC----------------------- -.((((.((((-----....))))))-))......----..(((..(((((.(.(--(((((((.(....).))))))))).)))))...)))..----------------------- ( -24.90, z-score = -1.81, R) >droWil1.scaffold_181089 6414890 114 + 12369635 -UGCAAAUGAUUUUCAAUAUAUCAUGUGUGUGAUACAUAUGUAUGUAGUUCGGUG--UUCUCCUUCA-AUCAAGAAGAAGUGGAACUUGUGACCCUUUAAUGUUUGUAUGUUCUUUUU -(((((((.........(((((((((((((...)))))))).)))))((((((.(--(((..((((.-........))))..))))))).)))........))))))).......... ( -23.40, z-score = -1.13, R) >droVir3.scaffold_12822 1097576 101 + 4096053 AUGUGAUCGAUUU---GUAGCUCAUAUAUGUUAAA------UAUGUAGUUCUGUG--CUCUCCUGCCUGUCGAGGAGAAGCGGAACUUGUGACCCUUUAAUGUAGA-AUAUUG----- .(((((.(.....---...).)))))(((((((((------.....((((((((.--.((((((........)))))).))))))))........)))))))))..-......----- ( -25.12, z-score = -2.16, R) >droMoj3.scaffold_6540 7399916 101 - 34148556 AUGUGAUUGGUUU---AUAGCUCAUAUAUGAUAUA------UAUGCAGUUCUUUG--CUCUCCUGCCUUUCGAGGAGAUGCGGAACUUGUGACCCUUUAAUGUAGA-AUAUUG----- ((((....((.((---(((((..(((((...))))------)..))((((((..(--(((((((........)))))).))))))))))))).))....))))...-......----- ( -22.40, z-score = -0.94, R) >droGri2.scaffold_14906 9634535 99 + 14172833 AUGUAA---AUUU---AUAGCUCAUAUAUGAUAAA------UAUGUAGUUCAGUGCUCUCUCCUACCUGUCGAGGAGAAGCGGAACUUGUGACCCUUUAAUGUUUU-AUAUG------ ((((((---(.((---(.((.((((((((.....)------)))))(((((...(((.((((((........))))))))).)))))...))..)).)))...)))-)))).------ ( -21.00, z-score = -1.45, R) >consensus _UGCAAAUGAU_____GUAAAUUAUC_GUGUGAUA________UAUAGUUCGGUG__UUCUCCCUCG_UUCAGGGAGAAUCGGAACUUGUGACCCAUUAAUGCAUA_AUAUU______ ..............................................(((((.(.....((((((........))))))..).)))))............................... (-12.95 = -12.42 + -0.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:39:50 2011