| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,953,541 – 19,953,636 |
| Length | 95 |
| Max. P | 0.990428 |

| Location | 19,953,541 – 19,953,636 |
|---|---|
| Length | 95 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 71.33 |
| Shannon entropy | 0.55607 |
| G+C content | 0.39924 |
| Mean single sequence MFE | -18.68 |
| Consensus MFE | -10.74 |
| Energy contribution | -10.78 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.990428 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
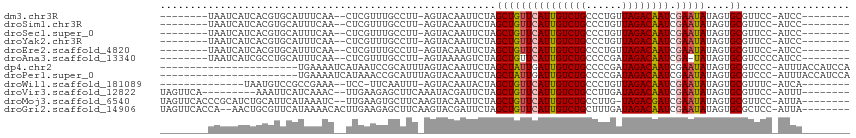
>dm3.chr3R 19953541 95 + 27905053 --------UAAUCAUCACGUGCAUUUCAA--CUCGUUUGCCUU-AGUACAAUUCUAGCUGUUCAUUGUCUGCCCUGUUAGACAAUCGAAUAUAGUGCGUUCC-AUCC-------- --------........(((..(.......--.......((..(-((.......)))))(((((((((((((......)))))))).)))))..)..)))...-....-------- ( -17.80, z-score = -1.83, R) >droSim1.chr3R 19784816 95 + 27517382 --------UAAUCAUCACGUGCAUUUCAA--CUCGUUUGCCUU-AGUACAAUUCUAGCUGUUCAUUGUCUGCCCUGUUAGACAAUCGAAUAUAGUGCGUUCC-AUCC-------- --------........(((..(.......--.......((..(-((.......)))))(((((((((((((......)))))))).)))))..)..)))...-....-------- ( -17.80, z-score = -1.83, R) >droSec1.super_0 20291583 95 + 21120651 --------UAAUCAUCACGUGCAUUUCAA--CUCGUUUGCCUU-AGUACAAUUCUAGCUGUUCAUUGUCUGCCCUGUUAGACAAUCGAAUAUAGUGCGUUCC-AUCC-------- --------........(((..(.......--.......((..(-((.......)))))(((((((((((((......)))))))).)))))..)..)))...-....-------- ( -17.80, z-score = -1.83, R) >droYak2.chr3R 2282517 95 - 28832112 --------UAAUCAUCACGUGCAUUUCAA--CUCGUUUGCCUU-AGUACAAUUCUAGCUGUUCAUUGUCUGCCCUGUUAGACAAUCGAAUAUAGUGCGUUCC-AUCC-------- --------........(((..(.......--.......((..(-((.......)))))(((((((((((((......)))))))).)))))..)..)))...-....-------- ( -17.80, z-score = -1.83, R) >droEre2.scaffold_4820 2364497 95 - 10470090 --------UAAUCAUCACGUGCAUUUCAA--CUCGUUUGCCUU-AGUACAAUUCUAGCUGUUCAUUGUCUGCCCUGUUAGACAAUCGAAUAUAGUGCGUUCC-AUCC-------- --------........(((..(.......--.......((..(-((.......)))))(((((((((((((......)))))))).)))))..)..)))...-....-------- ( -17.80, z-score = -1.83, R) >droAna3.scaffold_13340 1154879 95 - 23697760 --------UAAUCAUCGCCUGCAUUUCAA--CUCGUUUGCCUU-AGUAAAAGUCUAGCUGUUCAUUGUCUGCCCCGAUAGACAAUCGA-UAUAGUGCGUCCCCAUCC-------- --------.......(((...........--....(((((...-.)))))......(((((((((((((((......)))))))).))-.)))))))).........-------- ( -15.70, z-score = -1.24, R) >dp4.chr2 14486226 91 + 30794189 -----------------------UGAAAAUCAUAAUCCGCAUUUAGUACAAUUCUAGCUAUUGAUUGUCUGCCCCGAUAGACAAUCGAAUAUAGUGCGUCCC-AUUUACCAUCCA -----------------------..............((((((((((.........))))(((((((((((......)))))))))))....))))))....-............ ( -18.50, z-score = -3.13, R) >droPer1.super_0 5887284 91 - 11822988 -----------------------UGAAAAUCAUAAACCGCAUUUAGUACAAUUCUAGCUAUUGAUUGUCUGCCCCGAUAGACAAUCGAAUAUAGUGCGUCCC-AUUUACCAUCCA -----------------------..............((((((((((.........))))(((((((((((......)))))))))))....))))))....-............ ( -18.50, z-score = -3.28, R) >droWil1.scaffold_181089 6414753 88 - 12369635 --------------UAAUGUCCGCCGAAA--UCC-UUCAAUUU-AGUACAAUACUAGCUGUUCAUUGUCUGCCCUGUUAGACAAUCGAAUAUAGUGCGUUUC-AUCA-------- --------------..(((..(((.(((.--...-))).....-........((((..(((((((((((((......)))))))).))))))))))))...)-))..-------- ( -16.50, z-score = -1.94, R) >droVir3.scaffold_12822 1097416 95 - 4096053 UAGUUCA---------AAAUUCAUCAAAC--UUGAAGAGCUUCAAAUACGAUUCUAGCUGUUCAUUGUCUGCCUUGAUAGACAAUCGAAUAUAGUGCGUUCC-AUUU-------- .(((((.---------...((((......--.)))))))))......(((...(((..(((((((((((((......)))))))).))))))))..)))...-....-------- ( -17.90, z-score = -1.00, R) >droMoj3.scaffold_6540 7399754 103 + 34148556 UAGUUCACCCGCAUCUGCAUUCAUAAAUC--UUGAAGUGCUUCAAGUACAAUUCUAGCUGUUCAUUGUCUGCCUUG-UAGACGAUCGAAUAUAGUGCGUUCC-AUUA-------- .........(((((..((.((((......--.))))((((.....)))).......))((((((((((((((...)-)))))))).)))))..)))))....-....-------- ( -24.80, z-score = -2.48, R) >droGri2.scaffold_14906 9634378 104 - 14172833 UAGUUCACCA--AACUGCGUUCAUAAAACACUUGAAGAGCUUCAAGUACGAUUCUAGCUGUUCAUUGUCUGCUUUGAUAGACAAUCGAAUAUAGUGCGCUCC-AUUA-------- .((((.....--))))((((.(.......(((((((....)))))))...........(((((((((((((......)))))))).)))))..).))))...-....-------- ( -23.30, z-score = -1.28, R) >consensus ________UAAUCAUCACGUGCAUUUAAA__CUCGUUUGCCUU_AGUACAAUUCUAGCUGUUCAUUGUCUGCCCUGUUAGACAAUCGAAUAUAGUGCGUUCC_AUCC________ ........................................................(((((((((((((((......)))))))).)))))....)).................. (-10.74 = -10.78 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:39:48 2011