| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,950,143 – 19,950,216 |
| Length | 73 |
| Max. P | 0.645205 |

| Location | 19,950,143 – 19,950,216 |
|---|---|
| Length | 73 |
| Sequences | 11 |
| Columns | 77 |
| Reading direction | reverse |
| Mean pairwise identity | 60.25 |
| Shannon entropy | 0.83594 |
| G+C content | 0.44655 |
| Mean single sequence MFE | -13.78 |
| Consensus MFE | -2.95 |
| Energy contribution | -2.93 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.21 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.645205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 19950143 73 - 27905053 UUUGGAAAUUUAAUUAAAUAUACUAUCACUCACUUGUGCUCCUCCUCCUUGGUGCGCAUC----UGCACCAUCUCGU ...(((.((((....)))).......(((......))).))).......(((((((....----)))))))...... ( -11.80, z-score = -1.89, R) >droSec1.super_0 20288200 70 - 21120651 UUGAGAAAUUUAAUCACUU-UA--AUUACUCACUUGUGCUCCUCCUCCUUGGUGCGCAUC----UGCACCAUCUCGU ..((((...((((.....)-))--).....((..(((((.((........)).)))))..----)).....)))).. ( -14.10, z-score = -2.95, R) >droYak2.chr3R 2279153 73 + 28832112 UCUGGAAAUGUAAAUGUUUAUAGUAUCACCCACUUGUGCUCCUCCUCCUUGGUGCGCAUC----UGCACCAUCUCGU ..(((...(((((....)))))........((..(((((.((........)).)))))..----))..)))...... ( -13.30, z-score = -1.01, R) >droEre2.scaffold_4820 2361066 73 + 10470090 UCUAGAAAUUGCAUUAUUUAUAAUAGCACCCACUUGUGCUCCUCCUCCUUGGUGCGCAUC----UGCACCAUCUCGU ...(((..................(((((......))))).........(((((((....----))))))))))... ( -14.70, z-score = -2.12, R) >droAna3.scaffold_13340 1151550 62 + 23697760 --------UAUGGAUAAU--UAAUGU-ACUUACUUGUGCUCCUCUUCCUUGGUACGCAUU----AGUACUAUAUCGU --------....((((..--....((-(((....((((..((........))..))))..----)))))..)))).. ( -12.50, z-score = -1.95, R) >dp4.chr2 14482786 66 - 30794189 ------UAAGGACAGGUCUUUGCAGU-ACUCACUGGUGCUCCUCAUCCCUGGUGCGCAUU----UGUACCAUCUCAU ------...((((((((.....((((-....))))((((.((........)).)))))))----))).))....... ( -17.50, z-score = -1.04, R) >droWil1.scaffold_181089 6411462 64 + 12369635 --------CUCUUAUAUUCAAUUUGA-ACUCACCUGUGUUCUUCUUCGCGGGUUCGCAUG----AGCACGAUCUCAU --------...............(((-..(((((((((........)))))))..((...----.))..))..))). ( -12.30, z-score = -1.30, R) >droVir3.scaffold_12822 1093593 59 + 4096053 ----------CCUGGAU---GCCUGC-GCUUACUUGUGCUCCUCGUCCCGGGUGCGCAUG----UGCACAAUCUCAU ----------((.((((---(...((-((......))))....))))).))(((((....----)))))........ ( -21.30, z-score = -2.49, R) >droMoj3.scaffold_6540 7396155 55 - 34148556 ----------------CUGGGACAGU--CUUACUUGUGCUCCUCCUCUCUGGUGCGCAUU----UGCACAAUCUCAU ----------------.(((((..((--......(((((.((........)).)))))..----...))..))))). ( -14.30, z-score = -1.92, R) >droGri2.scaffold_14906 9630883 64 + 14172833 --------AUCGGACAAUCAUAAUUC-ACUUACUUGUGCUCCUCCUCUUUGGUGCGCAUG----UGCACAAUCUCAU --------..................-...(((.(((((.((........)).))))).)----))........... ( -11.70, z-score = -1.40, R) >apiMel3.Group16 4394848 62 - 5631066 ---------------AUUCAUCACGUAAUUGAUUUAUAUUAUUACGCUCUUGUUCAGUUUUUACUGCAGCUUCUCGU ---------------........((((((.(((....))))))))).....(((((((....)))).)))....... ( -8.10, z-score = -1.48, R) >consensus ________UUUAAAUAUUUAUAAUGU_ACUCACUUGUGCUCCUCCUCCUUGGUGCGCAUC____UGCACCAUCUCGU ...................................((((.((........))...)))).................. ( -2.95 = -2.93 + -0.02)

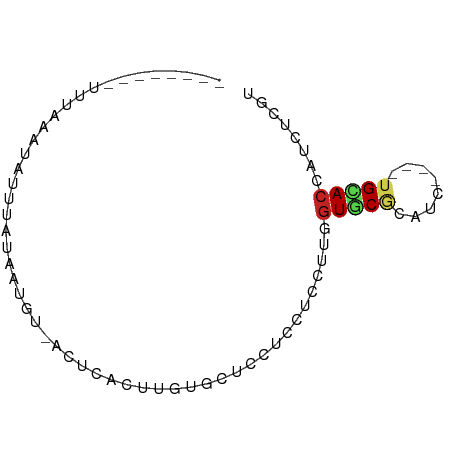
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:39:46 2011