
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,945,989 – 19,946,067 |
| Length | 78 |
| Max. P | 0.967864 |

| Location | 19,945,989 – 19,946,067 |
|---|---|
| Length | 78 |
| Sequences | 11 |
| Columns | 85 |
| Reading direction | forward |
| Mean pairwise identity | 71.54 |
| Shannon entropy | 0.60253 |
| G+C content | 0.52536 |
| Mean single sequence MFE | -11.50 |
| Consensus MFE | -10.25 |
| Energy contribution | -10.35 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.10 |
| Mean z-score | -0.89 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.967864 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 19945989 78 + 27905053 ------CUCACUCUCUCCAUCUUA-ACCAACUCCUUCCACAGAUGCAGGCGUAUCGCGAUAACUUCAAGCAAACGCCCUGCCCAU ------..................-...................((((((((...((...........))..))).))))).... ( -11.90, z-score = -1.84, R) >droAna3.scaffold_13340 1146996 76 - 23697760 UUCAUUCGCUCGCUUUCCAUUUCCCAUCGCCCCCACCCACAGAUGCAGGCGUACCGCGAUAACUUCAAGCAAACGC--------- .....((((.(((((..(((((..................))))).)))))....)))).................--------- ( -9.77, z-score = 0.14, R) >droEre2.scaffold_4820 2356942 76 - 10470090 --------CUCACUCUCUAUCUCA-ACCAACUCCUUUCGCAGAUGCAGGCGUAUCGCGAUAACUUCAAGCAAACGCCCUGCCCAU --------................-.............((((.....(((((...((...........))..))))))))).... ( -12.00, z-score = -1.10, R) >droYak2.chr3R 2275100 62 - 28832112 ----------------------CA-ACCAACUCCUUUCGCAGAUGCAGGCGUAUCGCGAUAACUUCAAGCAAACGCCCUGCCCAU ----------------------..-.............((((.....(((((...((...........))..))))))))).... ( -12.00, z-score = -1.02, R) >droSec1.super_0 20284056 74 + 21120651 ----------CUCACUCCAUCUCA-ACCAACUCCUUCCACAGAUGCAGGCGUAUCGCGAUAACUUCAAGCAAACGCCCUGCCCAU ----------..............-...................((((((((...((...........))..))).))))).... ( -11.90, z-score = -1.83, R) >droSim1.chr3R 19777637 78 + 27517382 ------CUCACUCGCUCCAUCUCA-ACCAACUCCUUCCACAGAUGCAGGCGUAUCGCGAUAACUUCAAGCAAACGCCCUGCCCAU ------..................-...................((((((((...((...........))..))).))))).... ( -11.90, z-score = -1.19, R) >droWil1.scaffold_181089 6407483 75 - 12369635 --------GUCACUUCUCCCUCU--UUCUCUCUUUGGCUCAGAUGCAGGCGUAUCGCGAUAACUUCAAGCAAACGCCCUGCCCAG --------...............--.........(((.......((((((((...((...........))..))).)))))))). ( -12.41, z-score = 0.07, R) >dp4.chr2 14478303 68 + 30794189 ---------------AUCAGUUU--UCGCUCCCAAUCCACAGAUGCAGGCGUAUCGCGAUAACUUCAAGCAAACGCCCUGCCCAU ---------------........--...................((((((((...((...........))..))).))))).... ( -11.90, z-score = -0.66, R) >droPer1.super_0 5879308 67 - 11822988 ---------------AUCAGUUU--UCGCUCCCAAUCCACAGAUGCAGGCGUAUCGCGAUAACUUCAAGCAAACGCCCUGCCCA- ---------------........--...................((((((((...((...........))..))).)))))...- ( -11.90, z-score = -0.63, R) >droVir3.scaffold_12822 1087282 78 - 4096053 ------UAUCCUCUACCCCUUUUGGUCCACCCCUUCCU-CAGAUGCAAGCGUAUCGCGAUAACUUCAAGCAAACGCCUUGCCCGU ------........(((......)))............-.....((((((((...((...........))..))).))))).... ( -10.20, z-score = -0.37, R) >droGri2.scaffold_14906 9625827 77 - 14172833 ------UAUCCUUUAACCCGUUC--UCAACCCCUGCCCACAGAUGCAAGCGUAUCGCGAUAACUUCAAGCAAACGCCUUGCCCAA ------.................--.......(((....)))..((((((((...((...........))..))).))))).... ( -10.60, z-score = -1.35, R) >consensus __________CUCUCUCCAUUUCA_ACCAACCCCUUCCACAGAUGCAGGCGUAUCGCGAUAACUUCAAGCAAACGCCCUGCCCAU ............................................((((((((...((...........))..)))).)))).... (-10.25 = -10.35 + 0.10)
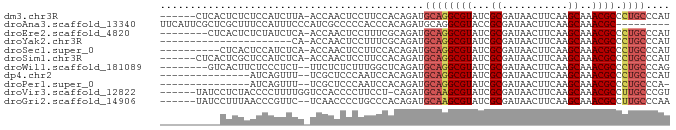

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:39:43 2011