
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,930,077 – 19,930,133 |
| Length | 56 |
| Max. P | 0.912533 |

| Location | 19,930,077 – 19,930,133 |
|---|---|
| Length | 56 |
| Sequences | 14 |
| Columns | 59 |
| Reading direction | reverse |
| Mean pairwise identity | 80.29 |
| Shannon entropy | 0.42088 |
| G+C content | 0.62333 |
| Mean single sequence MFE | -20.95 |
| Consensus MFE | -13.44 |
| Energy contribution | -13.42 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.912533 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 19930077 56 - 27905053 CGCCCAAUCCGAUGACGAGGA---GGAGACGGAGGUGGCCGUCAAUGUGGAUGGCCAUG ...((..(((........)))---(....)))..(((((((((......))))))))). ( -22.00, z-score = -2.21, R) >apiMel3.Group11 12411881 58 + 12576330 -GCCCAGUCGGACGAUGAUGACGUGGCGGACGAUGUGUCCGUCAAUGUUGGCGGGCCCG -((((.(((....)))(..((((((((((((.....))))))).)))))..)))))... ( -24.00, z-score = -1.15, R) >anoGam1.chr2L 48548990 56 - 48795086 -GCACAGAGCGAUGAUGAGGACAUGCCCGAAGAUGUGGCUGUCCCGGUGGAGGGCAG-- -.((((...((...(((....)))...))....))))(((.(((....))).)))..-- ( -13.60, z-score = 0.69, R) >droGri2.scaffold_14906 9607160 56 + 14172833 CGCCCAGUCGGACGAUGAGGA---AUCGGAGGAGGUGGCCGUCAAUGUGGAUGGCCAUG .(((...((...((((.....---))))..)).)))(((((((......)))))))... ( -20.00, z-score = -1.30, R) >droMoj3.scaffold_6540 7372797 56 - 34148556 CGCCCAGUCGGACGACGAGGA---GUCGGAAGAUGUGGCCGUCAAUGUGGAUGGCCAUG ..((..(((....)))..)).---(((....)))(((((((((......))))))))). ( -24.40, z-score = -2.89, R) >droVir3.scaffold_12822 1069207 56 + 4096053 CGCGCAGUCGGACGAUGAGGA---GUCGGAGGAUGUGGCCGUCAAUGUGGAUGGCCAUG .......((.(((........---))).))....(((((((((......))))))))). ( -20.60, z-score = -1.79, R) >droWil1.scaffold_181089 6390578 56 + 12369635 CGCCCAAUCGGAUGACGAGGA---AUCCGAGGAUGUGGCCGUCAAUGUUGAUGGCCAUG ...((..((((((........---))))))))..((((((((((....)))))))))). ( -24.70, z-score = -3.32, R) >droPer1.super_0 5860280 56 + 11822988 CGCCCAAUCGGACGAUGAGGA---AUCGGAAGAGGUGGCGGUCAAUGUGGAUGGACACG ((((...((...((((.....---))))...))...))))......(((......))). ( -16.10, z-score = -0.99, R) >dp4.chr2 14459832 56 - 30794189 CGCCCAAUCGGACGAUGAGGA---AUCGGAAGAGGUGGCGGUCAAUGUGGAUGGCCACG ..((..(((....)))..)).---.((....)).(((((.(((......))).))))). ( -19.70, z-score = -1.71, R) >droAna3.scaffold_12911 2141214 56 + 5364042 UGCCCAGUCCGACGACGAGGA---GGAGACCGAGGUGGCCGUCAACGUCGAUGGCCAUG ..((..(((....)))..)).---(....)....(((((((((......))))))))). ( -20.50, z-score = -1.14, R) >droEre2.scaffold_4820 2340955 56 + 10470090 CGCCCAAUCGGAUGACGAGGA---GGAGACGGAGGUGGCCGUCAAUGUGGAUGGCCAUG ...((..(((.....)))...---(....)))..(((((((((......))))))))). ( -21.30, z-score = -2.14, R) >droYak2.chr3R 2259248 56 + 28832112 CGCCCAAUCCGAUGACGAGGA---GGAGACGGAGGUGGCCGUCAAUGUGGAUGGCCAUG ...((..(((........)))---(....)))..(((((((((......))))))))). ( -22.00, z-score = -2.21, R) >droSec1.super_0 20268123 56 - 21120651 CGCCCAAUCCGACGACGAGGA---GGAGACGGAGGUGGCCGUCAAUGUGGAUGGCCAUG (.((...(((..(.....)..---)))...)).)(((((((((......))))))))). ( -22.40, z-score = -2.16, R) >droSim1.chr3R 19761657 56 - 27517382 CGCCCAAUCCGAUGACGAGGA---GGAGACGGAGGUGGCCGUCAAUGUGGAUGGCCAUG ...((..(((........)))---(....)))..(((((((((......))))))))). ( -22.00, z-score = -2.21, R) >consensus CGCCCAAUCGGACGACGAGGA___GGCGGAGGAGGUGGCCGUCAAUGUGGAUGGCCAUG ..................................(((((((((......))))))))). (-13.44 = -13.42 + -0.02)

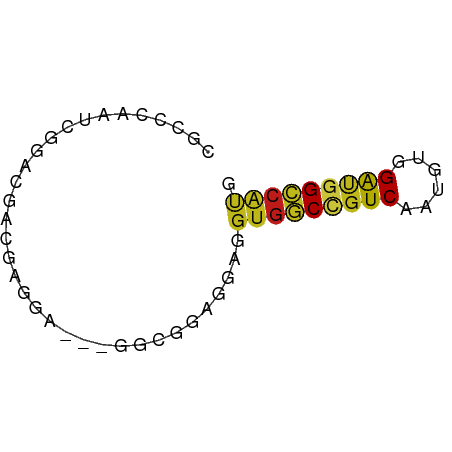
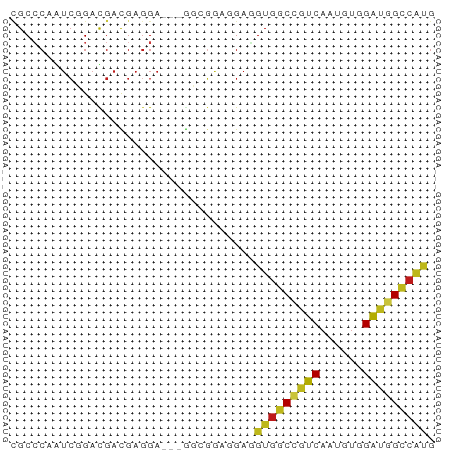
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:39:43 2011