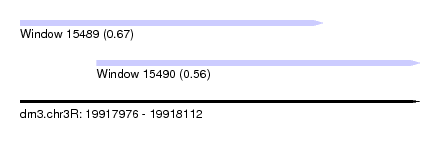
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,917,976 – 19,918,112 |
| Length | 136 |
| Max. P | 0.672241 |

| Location | 19,917,976 – 19,918,079 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 78.79 |
| Shannon entropy | 0.40295 |
| G+C content | 0.51230 |
| Mean single sequence MFE | -34.98 |
| Consensus MFE | -18.13 |
| Energy contribution | -18.63 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.672241 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 19917976 103 + 27905053 CGGAUAUUGAAGGGCAUG----GGUUACCAGCUUGGGGCUACGCCCACUCAUGUGAAAUCACCGCCCAGUUAAGCGCGGGCGAUUCAAAUAAAUUGGCCAACUUAAC ...........((((.((----(((....(((.....)))..))))).....(((....))).)))).((((((...((.(((((......))))).))..)))))) ( -29.90, z-score = -0.31, R) >droSim1.chr3R 19749569 103 + 27517382 CGGAAAUUGAAGGGCAUG----GGUAACCAGCUCGGGGUUACGCCCACCCAUGUGAAAUCACCGCCCAGUUAAGUGCGGGCGAUUCAAAUAAAUUGGCCAACUUAAC ...........(((((((----(((...(.....)(((.....)))))))))(((....))).)))).(((((((..((.(((((......))))).)).))))))) ( -35.00, z-score = -1.67, R) >droSec1.super_0 20256417 103 + 21120651 CGGAAAUUGAAGGGCAUG----GGUAACCAGCUCGGGGUUACGCCCACCCAUGUGAAAUCACCGCCCAGUUAAGUGCGGGCGAUUCAAAUAAAUUGGCCAACUUAAC ...........(((((((----(((...(.....)(((.....)))))))))(((....))).)))).(((((((..((.(((((......))))).)).))))))) ( -35.00, z-score = -1.67, R) >droYak2.chr3R 2246884 103 - 28832112 CGGAAAUUGAAGGGCAUG----GGUAGCUGGCUUGGGCUUACACCCAACCAUGCGAAAUCAACGCCCAGUUAAGUGCGGGCGAUUCACAUAAAUUGGCCAACUUAAC ...........((((...----(((.(((((.(((((......)))))))).))...)))...)))).(((((((..((.(((((......))))).)).))))))) ( -35.70, z-score = -2.46, R) >droEre2.scaffold_4820 2328623 103 - 10470090 CGGAAAUUGAAGGGCAUG----GGUAGCCAGCUUGGGGUUACAACCACCCAUGUGAAAACACCGCCCAGUUAAGUGCGGGCGAUUCACAUAAAUUGGCCAACUUAAC ...........(((((((----(((..((.....))(((....)))))))))((....))...)))).(((((((..((.(((((......))))).)).))))))) ( -35.10, z-score = -1.95, R) >droAna3.scaffold_12911 2129503 98 - 5364042 CAGCCAUGGGAGGGGGCGAAAUGGCAACCAUUUCUGGAGCA-GUCCACCC-UCUGGUAUC-UGAUAUAGUU------GGUCAGUUAAACGGAAUUGGCCAGCAUAGC (((..((.((((((...(((((((...)))))))((((...-.)))))))-))).))..)-)).....(((------((((((((......)))))))))))..... ( -39.20, z-score = -3.10, R) >consensus CGGAAAUUGAAGGGCAUG____GGUAACCAGCUCGGGGUUACGCCCACCCAUGUGAAAUCACCGCCCAGUUAAGUGCGGGCGAUUCAAAUAAAUUGGCCAACUUAAC ...........((((.......((...))......(((.....))).................)))).(((((((..((.(((((......))))).)).))))))) (-18.13 = -18.63 + 0.50)


| Location | 19,918,002 – 19,918,112 |
|---|---|
| Length | 110 |
| Sequences | 8 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 68.27 |
| Shannon entropy | 0.63934 |
| G+C content | 0.44802 |
| Mean single sequence MFE | -25.22 |
| Consensus MFE | -10.20 |
| Energy contribution | -9.64 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.561053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 19918002 110 + 27905053 GCUUGGGGCUACGCCCACUCAUGUGAAAUCACCGCCCAGUUAAGCGCGGGCGAUUCAAAUAAAUUGGCCAA-CUUAACUAGAAAAUAGCAGCAGUGAAUAGA---UUUCACAAA .....((((...)))).....(((((((((.......(((((((...((.(((((......))))).))..-)))))))........((....)).....))---))))))).. ( -29.50, z-score = -1.40, R) >droSim1.chr3R 19749595 110 + 27517382 GCUCGGGGUUACGCCCACCCAUGUGAAAUCACCGCCCAGUUAAGUGCGGGCGAUUCAAAUAAAUUGGCCAA-CUUAACUAGACAAUCGCAGCAGUGAACAGA---UUCCAUAAA .....(((.....))).....((((.((((.......((((((((..((.(((((......))))).)).)-)))))))......((((....))))...))---)).)))).. ( -29.10, z-score = -1.25, R) >droSec1.super_0 20256443 110 + 21120651 GCUCGGGGUUACGCCCACCCAUGUGAAAUCACCGCCCAGUUAAGUGCGGGCGAUUCAAAUAAAUUGGCCAA-CUUAACUAGACAAUCGCAGCAGUGAACAGA---UUCCAUAAA .....(((.....))).....((((.((((.......((((((((..((.(((((......))))).)).)-)))))))......((((....))))...))---)).)))).. ( -29.10, z-score = -1.25, R) >droYak2.chr3R 2246910 110 - 28832112 GCUUGGGCUUACACCCAACCAUGCGAAAUCAACGCCCAGUUAAGUGCGGGCGAUUCACAUAAAUUGGCCAA-CUUAACUAGACAAUCGCAGCAGUGAACAGA---UUCCAUAAA (.(((((......))))).)(((.(((.((.......((((((((..((.(((((......))))).)).)-)))))))......((((....))))...))---))))))... ( -27.90, z-score = -1.69, R) >droEre2.scaffold_4820 2328649 110 - 10470090 GCUUGGGGUUACAACCACCCAUGUGAAAACACCGCCCAGUUAAGUGCGGGCGAUUCACAUAAAUUGGCCAA-CUUAACUAGACAAUCGCAGCAGUGAACAGA---UUCCAUAAG ...(((((((.....(((....)))....(((.((..((((((((..((.(((((......))))).)).)-))))))).(.....)...)).)))....))---))))).... ( -26.60, z-score = -0.95, R) >droAna3.scaffold_12911 2129533 101 - 5364042 ---------UUUCUGGAGCAGUCCACCCUCUGGUAUCUGAUAUAGUUGGUCAGUUAAACGGAAUUGGCCAG-CAUAGCUGGAGAAACGCAGCGUGAAAUAAG--GUCCUGGAA- ---------....((((....))))...((..(.((((......(((((((((((......))))))))))-)...((((........))))........))--)).)..)).- ( -29.20, z-score = -0.71, R) >dp4.chr2 16348108 91 - 30794189 ---------------CACCCAUGUGAUAUACCAGCUCCGUUAAG--CUGGCGAUUCAAAUAAAUUGACAAAACUUAACUGGAGAAA---AUCCUUAAAAAAA---CCUCUGAAA ---------------(((....))).........((((((((((--.((.(((((......))))).))...)))))).))))...---.............---......... ( -13.50, z-score = -1.55, R) >droPer1.super_0 5976502 97 + 11822988 ---------------CACCCAUGUGAUAUACCAGCUCCGUUAAG--CUGGCGAUUCAAGUAAAUUGACAAAUCUUAACUGGAUAAACGUAGCAGUAGAGAAAAUGCCUCAAAAA ---------------........(((....((((((......))--))))....)))......((((((..((((.((((...........)))).))))...))..))))... ( -16.90, z-score = -0.55, R) >consensus GCU_GGGGUUACGCCCACCCAUGUGAAAUCACCGCCCAGUUAAGUGCGGGCGAUUCAAAUAAAUUGGCCAA_CUUAACUAGACAAUCGCAGCAGUGAACAGA___UUCCAUAAA ......................................((((((...((.(((((......))))).))...)))))).................................... (-10.20 = -9.64 + -0.56)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:39:42 2011