
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,852,646 – 19,852,752 |
| Length | 106 |
| Max. P | 0.619448 |

| Location | 19,852,646 – 19,852,752 |
|---|---|
| Length | 106 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 69.05 |
| Shannon entropy | 0.63286 |
| G+C content | 0.33847 |
| Mean single sequence MFE | -17.58 |
| Consensus MFE | -6.63 |
| Energy contribution | -6.80 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.64 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.619448 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 19852646 106 + 27905053 GAAAACGAUGAGGAAAACAU--UCCUCGAUGUUUAUCAGAA-AUCGAAAUGCCGCAAAGC-AUUUCACAUUUAUUUAUAAACAAAGAGCUGGGCACUUAAAAUACAAUCA (((((((.(((((((....)--)))))).))))).((((..-.(((((((((......))-)))))...((((....))))....)).))))...............)). ( -22.80, z-score = -2.91, R) >droAna3.scaffold_13340 15996481 86 - 23697760 CGAAAUGAUGAGGCUGAUGAGGCUCUUCGUGUUUAUCACAA-AUCGAAAUGGCACAUAUU--UUUGCAAUUUAUUUAUAAACACC-AGCA-------------------- ......((.(((.((....)).))).))((((((((...((-((..((((.(((......--..))).)))))))))))))))).-....-------------------- ( -15.40, z-score = -0.08, R) >droEre2.scaffold_4820 2265654 107 - 10470090 GAAAACGAUGAGGAAAACAU--UCCUGAAUGUUUAUCAGAA-AUCGAAAUACCGCAAAGCCAUUUCACAUUUAUUUAUAAACAAAGAGCUCGACACUUAAAAUACAAUUA .....((((.(((((....)--)))).).(((((((...((-((.(((((..(.....)..)))))..))))....)))))))......))).................. ( -12.40, z-score = -0.72, R) >droYak2.chr3R 2183165 107 - 28832112 GAAAGCGAUGAGGAAAACAU--UCCUGCAUGUUUAUCAGAA-AUCAAAAUACCGCACAGCCAUUUCACAUUUAUUUAUAAACAAAGAGCUCGACACUUAAAAUACAAUUA ...(((.((((((((....)--)))).)))((((((...((-((..((((..(.....)..))))...))))....)))))).....))).................... ( -10.40, z-score = 0.23, R) >droSec1.super_0 20194218 106 + 21120651 GAAAACGAUGAGGAAAACAU--UCCUCAAUGUUUAUCACAA-AUCGAAAUGCCGCAAAGC-AUUUCAUAUUUAUUUAUAAACAAAGAGCUGGGCACUUAAAAUACAAUCA (((((((.(((((((....)--)))))).))))).)).((.-.(((((((((......))-)))))(((......))).......))..))................... ( -22.00, z-score = -3.28, R) >droSim1.chr3R 19685776 106 + 27517382 GAAAACGAUGAGGAAAACAU--UCCUCAAUGUUUAUCACAA-AUCGAAAUGCCGCAAAGC-AUUUCACAUUUAUUUAUAAACAAAAAGCUGGGCACUUAAAAUACAAUCA (((((((.(((((((....)--)))))).))))).))....-...(((((((......))-)))))............................................ ( -21.20, z-score = -3.38, R) >droGri2.scaffold_14906 9522434 102 - 14172833 -AACAUUGGGCGAAAAAGCUGCUGCUCCGAGUUUAUCAAAUUGUUGUCGCCAGGCGUUCUAAUUAUAAACCCA---AUGAAAAGAUGGCUAAGAGCAAUAUAAACA---- -....(((((((..........))).))))((((((...((((((.(.((((....(((..............---..)))....))))..).)))))))))))).---- ( -18.89, z-score = 0.58, R) >consensus GAAAACGAUGAGGAAAACAU__UCCUCAAUGUUUAUCACAA_AUCGAAAUGCCGCAAAGC_AUUUCACAUUUAUUUAUAAACAAAGAGCUGGGCACUUAAAAUACAAUCA ..(((((.((((((........)))))).)))))............................................................................ ( -6.63 = -6.80 + 0.17)

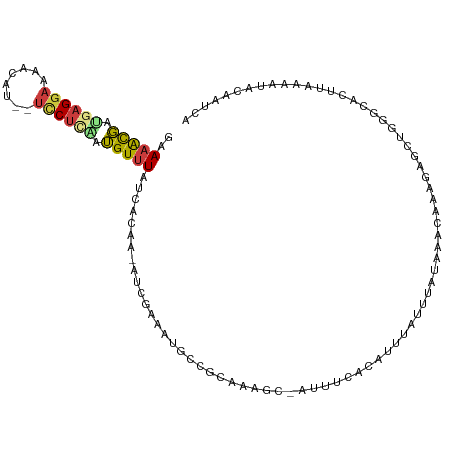
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:39:38 2011