
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,849,674 – 19,849,773 |
| Length | 99 |
| Max. P | 0.610560 |

| Location | 19,849,674 – 19,849,773 |
|---|---|
| Length | 99 |
| Sequences | 7 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 69.15 |
| Shannon entropy | 0.55061 |
| G+C content | 0.51938 |
| Mean single sequence MFE | -27.35 |
| Consensus MFE | -11.89 |
| Energy contribution | -11.49 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.610560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
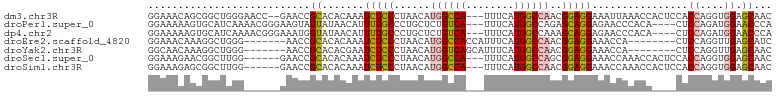
>dm3.chr3R 19849674 99 + 27905053 GGAAACAGCGGCUGGGAACC--GAACCGCACACAAAUCUCCCUAACAUGGCCA---UUUCAUGGCCAACGGAGGAAAUUAAACCACUCCACCAGGUGGAGCAAC ((.....((((.(.(....)--.).)))).......(((((......((((((---(...)))))))..)))))........)).((((((...)))))).... ( -31.50, z-score = -2.60, R) >droPer1.super_0 6054461 97 + 11822988 GGAAAAAGUGCAUCAAAACGGGAAGUAGUAUAACAUUUUGCCCUGCUCUGUCA---UUUCAUGGCCAGAGCAGGAGAACCCACA----CUCCAGAUGGAACCCA ((.................(((.....(((........)))((((((((((((---(...)))).)))))))))....)))...----.(((....)))..)). ( -27.90, z-score = -1.69, R) >dp4.chr2 16420509 97 - 30794189 GGAAAAAGUGCAUCAAAACGGGAAAUGGUAUAACAUUUUGCCCUGCUCUGUCA---UUUCAUGGCCAAAGCAGGAGAACCCACA----CUCCAGAUGGAACCCA ((.....(((..((.....(.((((((......)))))).)((((((..((((---(...)))))...)))))).))...))).----.(((....)))..)). ( -24.80, z-score = -0.60, R) >droEre2.scaffold_4820 2262682 89 - 10470090 GGAAACAAAGGCUGGG-------AACCGCACACAAAUCUCCCUAACAUGGCCACCAUUUCAUGGCCAACGGAGGAAACCA--------CUCCAGGUUGAGCAUC (....)....((((((-------(..............))))((((.((((((........))))))..((((.......--------))))..)))))))... ( -25.74, z-score = -1.57, R) >droYak2.chr3R 2180069 89 - 28832112 GGCAACAAAGGCUGGG-------AACCGCACACGAAUCUCCCUAACAUGGUCAGCAUUUCAUGGCCAACGGAGGAAACCA--------CUCCAGGUUGAGCAAC (....)....((((((-------(..((....))....))))((((.((((((........))))))..((((.......--------))))..)))))))... ( -24.10, z-score = -0.76, R) >droSec1.super_0 20191298 95 + 21120651 GGAAAGAACGGCUUGG------GAACCGCACACAAAUCUCCCUAACAUGGCCA---UUUCAUGGCCAGCGGAGGAAACCAAACCACUCCACCAGGUGGAGCAAC .........((.((((------.....(....)...(((((......((((((---(...)))))))..)))))...)))).)).((((((...)))))).... ( -27.60, z-score = -1.15, R) >droSim1.chr3R 19682482 95 + 27517382 GGAAAGAGCGGCUUGG------GAACCGCACACAAAUCUCCCUAACAUGGCCA---UUUCAUGGCCAACGGAGGAAACCAAACCACUCCACCAGGUGGAGCAAC ((.....((((.....------...)))).......(((((......((((((---(...)))))))..)))))...))......((((((...)))))).... ( -29.80, z-score = -1.86, R) >consensus GGAAACAGCGGCUGGG______GAACCGCACACAAAUCUCCCUAACAUGGCCA___UUUCAUGGCCAACGGAGGAAACCAAACC____CUCCAGGUGGAGCAAC ...........................((.......(((((......((((((........))))))..)))))................((....)).))... (-11.89 = -11.49 + -0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:39:37 2011