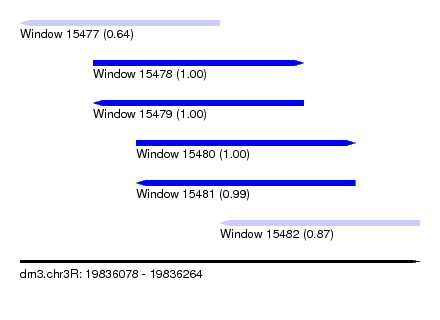
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,836,078 – 19,836,264 |
| Length | 186 |
| Max. P | 0.998855 |

| Location | 19,836,078 – 19,836,171 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 83.21 |
| Shannon entropy | 0.30424 |
| G+C content | 0.36921 |
| Mean single sequence MFE | -19.36 |
| Consensus MFE | -14.48 |
| Energy contribution | -15.24 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.641946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
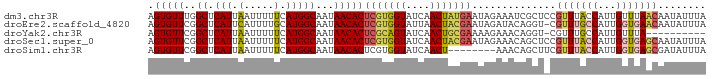
>dm3.chr3R 19836078 93 - 27905053 AGUGUUUGGCUCAUUAAUUUUUCAUGGCAAUAACACUCGUGGUAUCAACUAUGAAUAGAAAUCGCUCCGUUUACCAUUGUUUAACAAUAUUUA .(((((..((.(((.........)))))...)))))(((((((....))))))).....................((((.....))))..... ( -14.10, z-score = -0.18, R) >droEre2.scaffold_4820 2249106 92 + 10470090 AGUGUUCGGCUCAUUCAUUUUGCAUGGCAAUAACACUCGUGGUAUUAACUACGAAUAGAUACAGGU-CGUUUGCCAUUGGUGAACAAUAUUUA .(((((..((.(((.(.....).)))))...)))))(((((((....)))))))...(((....))-)(((..((...))..)))........ ( -23.00, z-score = -1.78, R) >droYak2.chr3R 2166526 82 + 28832112 AGUGUUCGGCUCAUUAAUUUUUCAUGGCAAUAACACUCGCAGUAUCAACUGCGAAAAGAAACAGGU-CGUUUGCCAUUGUUUA---------- .(((((..((.(((.........)))))...)))))(((((((....)))))))....((((((((-.....)))..))))).---------- ( -20.20, z-score = -2.01, R) >droSec1.super_0 20177592 93 - 21120651 AGUGUUCGGCUCAUUAAUUUUUCAUGGCAAUAACACUCGUGGUAUCAACUACGAAUAGAAACAGCUCCGUUUACCAUUGGUGAGCAAUAUUUA .(((((..((.(((.........)))))...)))))(((((((....)))))))..............(((((((...)))))))........ ( -21.50, z-score = -1.95, R) >droSim1.chr3R 19667003 85 - 27517382 AGUGUUCGGCUCAUUAAUUUUUCAUGGCAAUAACACUCGUGGUAUCAACU--------AAACAGCUUCGUUUACCAUUGGUGAGCGAUAUUUA ((((((..((.(((.........)))))...))))))..((((....)))--------).......(((((((((...)))))))))...... ( -18.00, z-score = -1.34, R) >consensus AGUGUUCGGCUCAUUAAUUUUUCAUGGCAAUAACACUCGUGGUAUCAACUACGAAUAGAAACAGCU_CGUUUACCAUUGGUGAACAAUAUUUA .(((((..((.(((.(.....).)))))...)))))(((((((....)))))))..............(((((((...)))))))........ (-14.48 = -15.24 + 0.76)
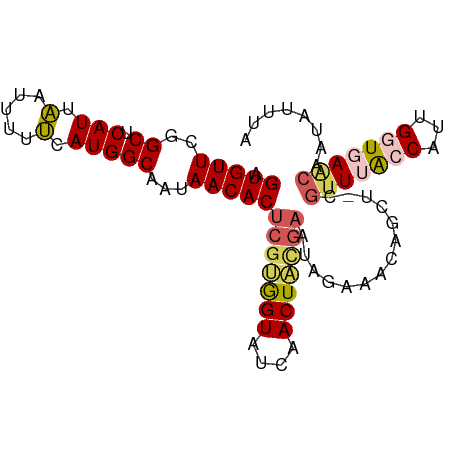
| Location | 19,836,112 – 19,836,210 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 91.26 |
| Shannon entropy | 0.15896 |
| G+C content | 0.33937 |
| Mean single sequence MFE | -21.30 |
| Consensus MFE | -21.10 |
| Energy contribution | -20.62 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.67 |
| Structure conservation index | 0.99 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.52 |
| SVM RNA-class probability | 0.998855 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
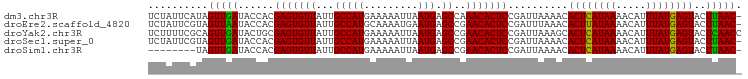
>dm3.chr3R 19836112 98 + 27905053 UCUAUUCAUAGUUGAUACCACGAGUGUUAUUGCCAUGAAAAAUUAAUGAGCCAAACACUCCGAUUAAAACACUCAUAAAACAUUUAUGAGUACUUAAC- ..........(((((......(((((((...(((((.........))).))..)))))))..........((((((((.....))))))))..)))))- ( -19.60, z-score = -3.48, R) >droEre2.scaffold_4820 2249139 98 - 10470090 UCUAUUCGUAGUUAAUACCACGAGUGUUAUUGCCAUGCAAAAUGAAUGAGCCGAACACUCCGAUUUAAACACUUAUAAAACAUUUAUGAGUACUUAAC- ..........(((((......(((((((...(((((.(.....).))).))..)))))))..........((((((((.....))))))))..)))))- ( -18.50, z-score = -2.41, R) >droYak2.chr3R 2166549 99 - 28832112 UCUUUUCGCAGUUGAUACUGCGAGUGUUAUUGCCAUGAAAAAUUAAUGAGCCGAACACUCCGAUUAAAGCACUCAUAAAACAUUUAUGAGUACUCAACC .....(((((((....)))))(((((((...(((((.........))).))..))))))).))....((.((((((((.....)))))))).))..... ( -27.00, z-score = -4.52, R) >droSec1.super_0 20177626 98 + 21120651 UCUAUUCGUAGUUGAUACCACGAGUGUUAUUGCCAUGAAAAAUUAAUGAGCCGAACACUCCGAUUAAAACACUCAUAAAACAUUUAUGAGUACUUAAC- ..........(((((......(((((((...(((((.........))).))..)))))))..........((((((((.....))))))))..)))))- ( -20.70, z-score = -3.28, R) >droSim1.chr3R 19667037 90 + 27517382 --------UAGUUGAUACCACGAGUGUUAUUGCCAUGAAAAAUUAAUGAGCCGAACACUCCGAUUAAAACACUCAUAAAACAUUUAUGAGUACUUAAC- --------..(((((......(((((((...(((((.........))).))..)))))))..........((((((((.....))))))))..)))))- ( -20.70, z-score = -4.64, R) >consensus UCUAUUCGUAGUUGAUACCACGAGUGUUAUUGCCAUGAAAAAUUAAUGAGCCGAACACUCCGAUUAAAACACUCAUAAAACAUUUAUGAGUACUUAAC_ ..........(((((......(((((((...(((((.........))).))..)))))))..........((((((((.....))))))))..))))). (-21.10 = -20.62 + -0.48)
| Location | 19,836,112 – 19,836,210 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 91.26 |
| Shannon entropy | 0.15896 |
| G+C content | 0.33937 |
| Mean single sequence MFE | -25.22 |
| Consensus MFE | -23.82 |
| Energy contribution | -22.98 |
| Covariance contribution | -0.84 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.99 |
| SVM RNA-class probability | 0.996826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
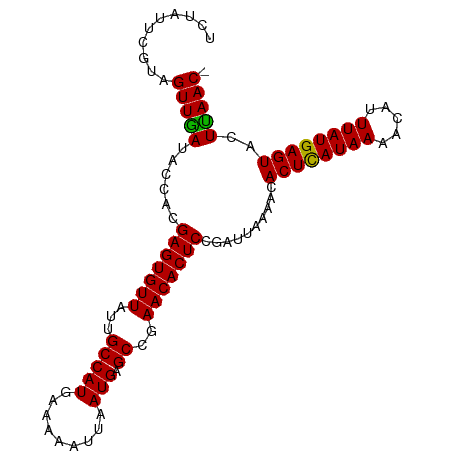
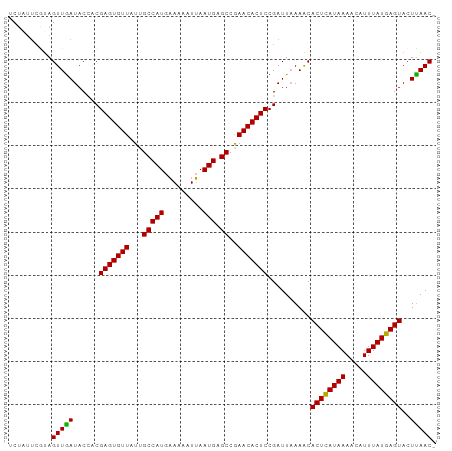
>dm3.chr3R 19836112 98 - 27905053 -GUUAAGUACUCAUAAAUGUUUUAUGAGUGUUUUAAUCGGAGUGUUUGGCUCAUUAAUUUUUCAUGGCAAUAACACUCGUGGUAUCAACUAUGAAUAGA -......(((((((((.....))))))))).((((.((.(((((((..((.(((.........)))))...)))))))(((((....))))))).)))) ( -23.10, z-score = -2.28, R) >droEre2.scaffold_4820 2249139 98 + 10470090 -GUUAAGUACUCAUAAAUGUUUUAUAAGUGUUUAAAUCGGAGUGUUCGGCUCAUUCAUUUUGCAUGGCAAUAACACUCGUGGUAUUAACUACGAAUAGA -.(((((((((.((((.....)))).))))))))).((.(((((((..((.(((.(.....).)))))...)))))))(((((....)))))))..... ( -22.20, z-score = -1.99, R) >droYak2.chr3R 2166549 99 + 28832112 GGUUGAGUACUCAUAAAUGUUUUAUGAGUGCUUUAAUCGGAGUGUUCGGCUCAUUAAUUUUUCAUGGCAAUAACACUCGCAGUAUCAACUGCGAAAAGA ((((((((((((((((.....)))))))))).)))))).(((((((..((.(((.........)))))...)))))))(((((....)))))....... ( -34.60, z-score = -5.25, R) >droSec1.super_0 20177626 98 - 21120651 -GUUAAGUACUCAUAAAUGUUUUAUGAGUGUUUUAAUCGGAGUGUUCGGCUCAUUAAUUUUUCAUGGCAAUAACACUCGUGGUAUCAACUACGAAUAGA -......(((((((((.....))))))))).((((.((.(((((((..((.(((.........)))))...)))))))(((((....))))))).)))) ( -25.00, z-score = -2.98, R) >droSim1.chr3R 19667037 90 - 27517382 -GUUAAGUACUCAUAAAUGUUUUAUGAGUGUUUUAAUCGGAGUGUUCGGCUCAUUAAUUUUUCAUGGCAAUAACACUCGUGGUAUCAACUA-------- -(((...(((((((((.....))))))))).....(((((((((((..((.(((.........)))))...))))))).))))...)))..-------- ( -21.20, z-score = -2.85, R) >consensus _GUUAAGUACUCAUAAAUGUUUUAUGAGUGUUUUAAUCGGAGUGUUCGGCUCAUUAAUUUUUCAUGGCAAUAACACUCGUGGUAUCAACUACGAAUAGA ....((((((((((((.....))))))))))))......(((((((..((.(((.(.....).)))))...)))))))(((((....)))))....... (-23.82 = -22.98 + -0.84)
| Location | 19,836,132 – 19,836,234 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 83.56 |
| Shannon entropy | 0.25694 |
| G+C content | 0.35307 |
| Mean single sequence MFE | -22.34 |
| Consensus MFE | -18.86 |
| Energy contribution | -18.67 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.50 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.32 |
| SVM RNA-class probability | 0.998312 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
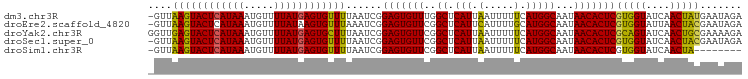
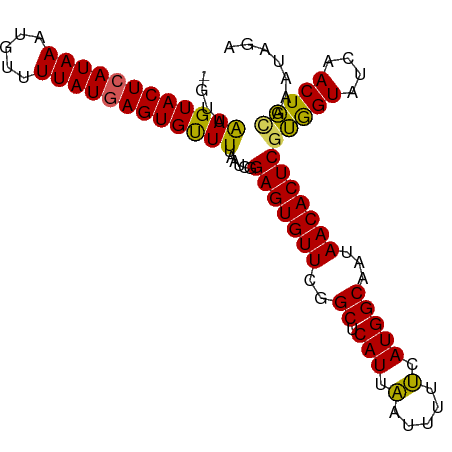
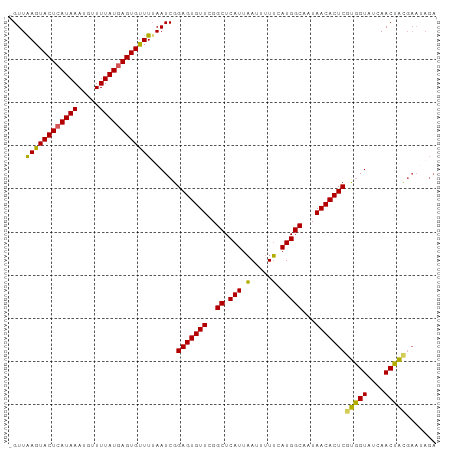
>dm3.chr3R 19836132 102 + 27905053 CGAGUGUUAUUGCCAUGAAAAAUUAAUGAGCCAAACACUCCGAUUAAAACACUCAUAAAACAUUUAUGAGUACUUAACC-GGCAGAACACAUAUUCAGUACAU----------- ...(((((.(((((........((((((((.......)))..)))))...((((((((.....))))))))........-)))))))))).............----------- ( -22.10, z-score = -3.84, R) >droEre2.scaffold_4820 2249159 108 - 10470090 CGAGUGUUAUUGCCAUGCAAAAUGAAUGAGCCGAACACUCCGAUUUAAACACUUAUAAAACAUUUAUGAGUACUUAACC-AGCAGAACAUAUACAUG-CCUAUU----UCGACG .(((((((...(((((.(.....).))).))..)))))))(((..((...((((((((.....))))))))........-.(((...........))-).))..----)))... ( -21.10, z-score = -3.01, R) >droYak2.chr3R 2166569 113 - 28832112 CGAGUGUUAUUGCCAUGAAAAAUUAAUGAGCCGAACACUCCGAUUAAAGCACUCAUAAAACAUUUAUGAGUACUCAACCCAGCAGAACACAUACAUA-CAUAUUCAUUUCGACG .(((((((...(((((.........))).))..)))))))(((....((.((((((((.....)))))))).))..........(((..........-....)))...)))... ( -20.24, z-score = -2.81, R) >droSec1.super_0 20177646 107 + 21120651 CGAGUGUUAUUGCCAUGAAAAAUUAAUGAGCCGAACACUCCGAUUAAAACACUCAUAAAACAUUUAUGAGUACUUAACC-GGCAGAACGUACAUAUA-UAUAUGUAUAU----- .(((((((...(((((.........))).))..)))))))((.((((...((((((((.....))))))))..)))).)-).......(((((((..-..)))))))..----- ( -25.90, z-score = -4.36, R) >consensus CGAGUGUUAUUGCCAUGAAAAAUUAAUGAGCCGAACACUCCGAUUAAAACACUCAUAAAACAUUUAUGAGUACUUAACC_AGCAGAACACAUACAUA_CAUAUU____U_____ .(((((((...(((((.........))).))..)))))))..........((((((((.....))))))))........................................... (-18.86 = -18.67 + -0.19)

| Location | 19,836,132 – 19,836,234 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 83.56 |
| Shannon entropy | 0.25694 |
| G+C content | 0.35307 |
| Mean single sequence MFE | -27.12 |
| Consensus MFE | -23.23 |
| Energy contribution | -22.72 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.989174 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
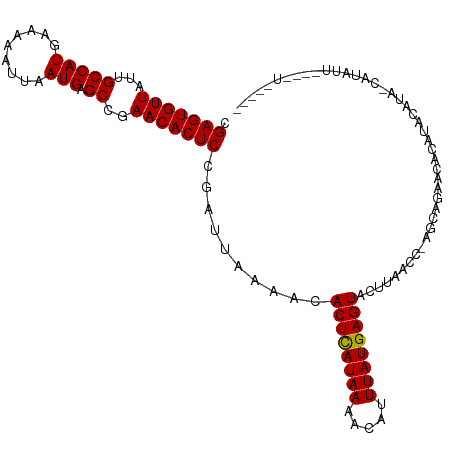
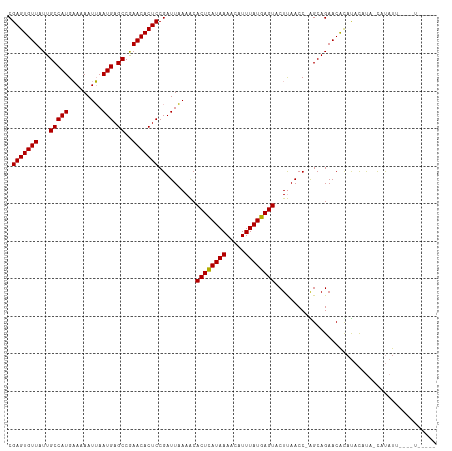
>dm3.chr3R 19836132 102 - 27905053 -----------AUGUACUGAAUAUGUGUUCUGCC-GGUUAAGUACUCAUAAAUGUUUUAUGAGUGUUUUAAUCGGAGUGUUUGGCUCAUUAAUUUUUCAUGGCAAUAACACUCG -----------.......((((....))))...(-((((((((((((((((.....))))))))).))))))))(((((((..((.(((.........)))))...))))))). ( -26.30, z-score = -2.75, R) >droEre2.scaffold_4820 2249159 108 + 10470090 CGUCGA----AAUAGG-CAUGUAUAUGUUCUGCU-GGUUAAGUACUCAUAAAUGUUUUAUAAGUGUUUAAAUCGGAGUGUUCGGCUCAUUCAUUUUGCAUGGCAAUAACACUCG ...(((----..((((-(((((.((((...((((-.....))))..)))).)).........)))))))..)))(((((((..((.(((.(.....).)))))...))))))). ( -22.20, z-score = -0.66, R) >droYak2.chr3R 2166569 113 + 28832112 CGUCGAAAUGAAUAUG-UAUGUAUGUGUUCUGCUGGGUUGAGUACUCAUAAAUGUUUUAUGAGUGCUUUAAUCGGAGUGUUCGGCUCAUUAAUUUUUCAUGGCAAUAACACUCG .........(((((((-(....)))))))).....((((((((((((((((.....)))))))))).)))))).(((((((..((.(((.........)))))...))))))). ( -32.20, z-score = -3.15, R) >droSec1.super_0 20177646 107 - 21120651 -----AUAUACAUAUA-UAUAUGUACGUUCUGCC-GGUUAAGUACUCAUAAAUGUUUUAUGAGUGUUUUAAUCGGAGUGUUCGGCUCAUUAAUUUUUCAUGGCAAUAACACUCG -----...((((((..-..))))))........(-((((((((((((((((.....))))))))).))))))))(((((((..((.(((.........)))))...))))))). ( -27.80, z-score = -3.46, R) >consensus _____A____AAUAUA_UAUAUAUAUGUUCUGCC_GGUUAAGUACUCAUAAAUGUUUUAUGAGUGUUUUAAUCGGAGUGUUCGGCUCAUUAAUUUUUCAUGGCAAUAACACUCG ...................................((((((((((((((((.....)))))))))))).)))).(((((((..((.(((.(.....).)))))...))))))). (-23.23 = -22.72 + -0.50)

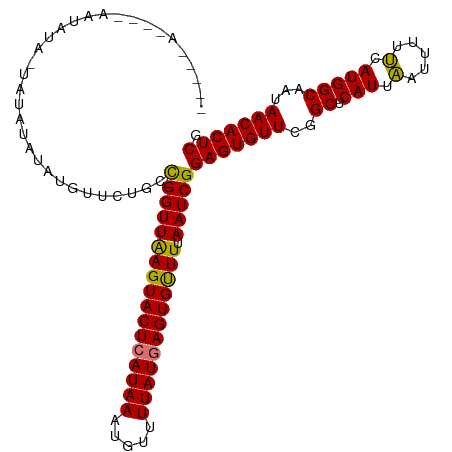
| Location | 19,836,171 – 19,836,264 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 55.97 |
| Shannon entropy | 0.79187 |
| G+C content | 0.34275 |
| Mean single sequence MFE | -21.24 |
| Consensus MFE | -4.14 |
| Energy contribution | -7.00 |
| Covariance contribution | 2.86 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.19 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.868129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 19836171 93 - 27905053 UUACGAAAAAUUC-AGAGGAUUGUAAGGAAUAUGUA-----C-----UGAAUAUGUGUUCUGCC-GGUUAAGUACUCAUAAAUGUUUUAUGAGUGUUUUAAUCGG ((((((.......-......))))))((((((((((-----.-----....)))))))))).((-((((((((((((((((.....))))))))).))))))))) ( -22.72, z-score = -2.74, R) >droEre2.scaffold_4820 2249198 99 + 10470090 UUCCGAAUAUUUC-AAAGGACGUAAUGGAAUCGUCGA----AAUAGGCAUGUAUAUGUUCUGCU-GGUUAAGUACUCAUAAAUGUUUUAUAAGUGUUUAAAUCGG ..((((.....((-(.((((((((.....((.(((..----....)))))...))))))))..)-))(((((((((.((((.....)))).))))))))).)))) ( -18.30, z-score = -0.73, R) >droYak2.chr3R 2166608 105 + 28832112 UUCCGAAAAUUUCGAAAGGACUGCAAGGAAUCGUCGAAAUGAAUAUGUAUGUAUGUGUUCUGCUGGGUUGAGUACUCAUAAAUGUUUUAUGAGUGCUUUAAUCGG ((((......(((....)))......))))..........((((((((....))))))))..((((...((((((((((((.....))))))))))))...)))) ( -28.40, z-score = -2.88, R) >droSec1.super_0 20177685 98 - 21120651 UUACGAAAAAUUC-AGAGGACUGUAAGGAAUAUAUA-----CAUAUAUAUAUGUACGUUCUGCC-GGUUAAGUACUCAUAAAUGUUUUAUGAGUGUUUUAAUCGG .............-..(((((.(((....(((((((-----....))))))).)))))))).((-((((((((((((((((.....))))))))).))))))))) ( -24.20, z-score = -2.74, R) >dp4.chr2 14353183 71 - 30794189 ---------UCUCUAUAACUCUAUGGAAACCAAUCAAUAACUACUCGCAUGUGCAGUCGCUUCUUAG--AAACUGCAAUCAA----------------------- ---------..((((((....))))))........................((((((..((....))--..)))))).....----------------------- ( -12.60, z-score = -2.70, R) >consensus UUACGAAAAUUUC_AAAGGACUGUAAGGAAUAGUCAA____AAUAUGUAUGUAUAUGUUCUGCU_GGUUAAGUACUCAUAAAUGUUUUAUGAGUGUUUUAAUCGG .................................................................((((((((((((((((.....)))))))))))).)))).. ( -4.14 = -7.00 + 2.86)
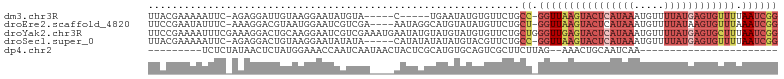

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:39:35 2011