
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,830,837 – 19,830,971 |
| Length | 134 |
| Max. P | 0.546823 |

| Location | 19,830,837 – 19,830,971 |
|---|---|
| Length | 134 |
| Sequences | 6 |
| Columns | 141 |
| Reading direction | forward |
| Mean pairwise identity | 64.54 |
| Shannon entropy | 0.67301 |
| G+C content | 0.35483 |
| Mean single sequence MFE | -30.72 |
| Consensus MFE | -10.09 |
| Energy contribution | -9.38 |
| Covariance contribution | -0.71 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.546823 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 19830837 134 + 27905053 AUGGAAAUGCCUUAUA----GAUUAGCUUAUGCUA---AUUUCUGGGAAAUUCCUGAAUAGGCAUGGGAAGAAGAAAUUAGAACAAGUUAAUCCUAAGAGAUAUCUGACAGAAUCUAUGCAAAUAUUCGAUAAUCCAAAUC .((((.....((((..----(((((((((...(((---((((((......(((((..........)))))..)))))))))...))))))))).)))).(((((.((.((.......)))).)))))......)))).... ( -30.10, z-score = -1.40, R) >droEre2.scaffold_4820 2243808 106 - 10470090 AUGGAAAUGCCUUAUA----GAUUAGCUUAUGCUA---AUUUCUGCGAAAUUCCUUAAAA-----GGGUAGAAGGAACUAGAACAAGGUAUUCCUAAGAGAUAUCUGACUGAAUCUAG----------------------- ..((((.((((((.((----(.(((((....))))---)(((((((......(((....)-----)))))))))...)))....))))))))))....((((.((.....))))))..----------------------- ( -24.10, z-score = -1.15, R) >droYak2.chr3R 2161201 118 - 28832112 AUGGAAAUGCCUUAUACAUAGAUUAGCUUAUGCUA---AUUUUUGGGAAAUUCCGGAAAAGGCAUGGAUGGAAGGCACUAGAACAAGGUAAUCCUGAAAGAUAUCAGACUGAAUCUAAAUA-------------------- ......(((((((...((.((((((((....))))---)))).))((.....))....)))))))((((...((((.((......))))....((((......)))).))..)))).....-------------------- ( -26.80, z-score = -1.43, R) >droSec1.super_0 20172327 134 + 21120651 AUGGAAAUGCCUUAUA----GAUUAGCUUAUGCUA---AUUUCUGGGAAAUUCCUAAAAAGGCAUGGCUAGAAGGAACUAGAACAAGGUAAUCCUAAGAGAUAUCUGACUGAAUCUAGUUAAAUAUUCGAUCAUCCAAAGC .((((.(((((((..(----(((((((....))))---)))).((((.....))))..))))))).....(((..(((((((...(((....))).(((....))).......))))))).....))).....)))).... ( -35.50, z-score = -2.41, R) >droSim1.chr3R 19660103 134 + 27517382 AUGGAAAUGCCUUAUA----GAUUAGCUUAUGCUA---AUUUCUGGGAAAUUCCUAAAAAGGCAUGGCUAGAAGGAACUAGAACAAGGUAAUCCUAAGAUAUAUCUCACUGAAUCUAGUUAAAUAUUCGAUCAUCCAACGC .((((.(((((((..(----(((((((....))))---)))).((((.....))))..))))))).....(((..(((((((...(((....)))........((.....)).))))))).....))).....)))).... ( -34.40, z-score = -2.50, R) >dp4.chr2 14348247 137 + 30794189 AGAGGGAGGACCUAUAGCACUGCUCCUAUGCACUGCCUAUAACUGAAGGUGCACUGUAAAAGGCUAUCCCUGAUGAUGUGGAUCAUGUAUAUACUUAGGUAUUAUGAUUCAAAUAUGUCUACUAUAUAGUACAUAGG---- ..(((((((.(((.(.(((.(((.(((...((.(.......).)).))).))).))).).))))).))))).......(((((((((...((((....)))))))))))))..((((((((.....))).)))))..---- ( -33.40, z-score = 0.34, R) >consensus AUGGAAAUGCCUUAUA____GAUUAGCUUAUGCUA___AUUUCUGGGAAAUUCCUGAAAAGGCAUGGCUAGAAGGAACUAGAACAAGGUAAUCCUAAGAGAUAUCUGACUGAAUCUAGUUAAAUAUUCGAUCAUCCA____ ..(((..((((((..........((((....)))).....((((((....(((.................)))....)))))).)))))).)))....((((.((.....))))))......................... (-10.09 = -9.38 + -0.71)

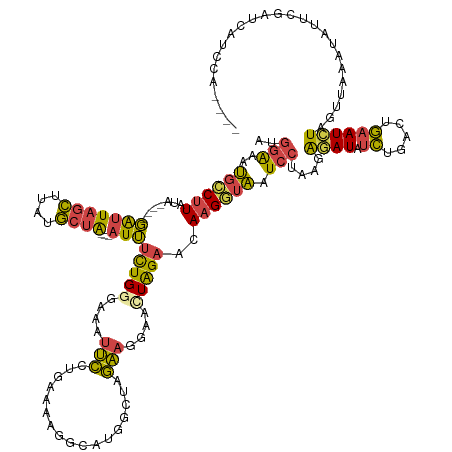
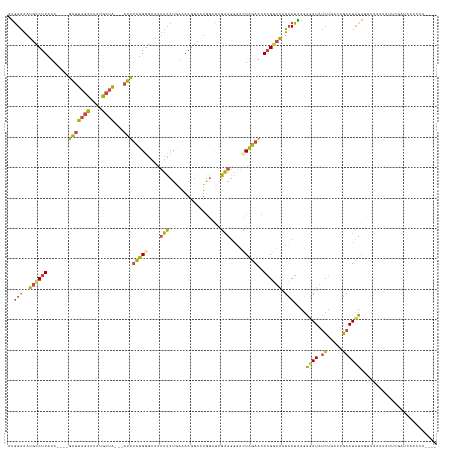
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:39:30 2011