| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,470,383 – 8,470,502 |
| Length | 119 |
| Max. P | 0.987682 |

| Location | 8,470,383 – 8,470,502 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 143 |
| Reading direction | forward |
| Mean pairwise identity | 70.43 |
| Shannon entropy | 0.46490 |
| G+C content | 0.50200 |
| Mean single sequence MFE | -42.67 |
| Consensus MFE | -22.55 |
| Energy contribution | -25.55 |
| Covariance contribution | 3.00 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.987682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
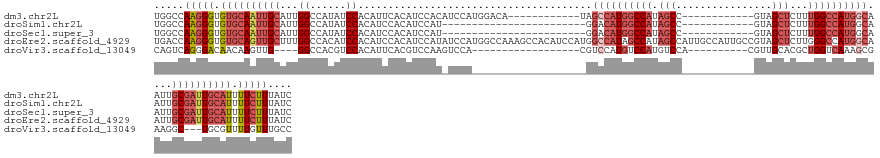
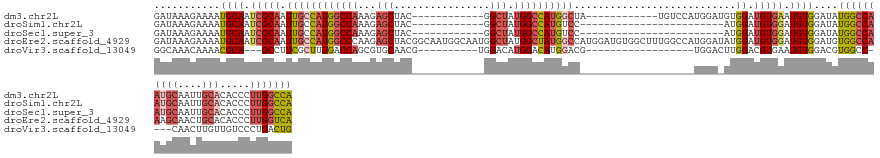
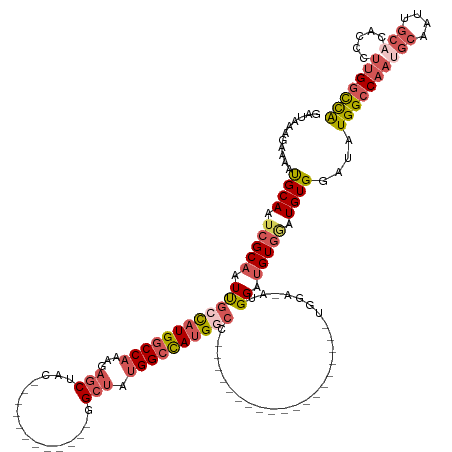
>dm3.chr2L 8470383 119 + 23011544 UGGCCAAGGGUGUGCAAUUGCAUUGGCCAUAUCCACAUUCACAUCCACAUCCAUGGACA------------UAGCCAUGGCCAUAGCC------------GUAGCUCUUUGGCCAUGGCAAUUGCGAUUGCAUUUUCUUUAUC .....(((((.(((((((((((.(((......)))........((((......))))..------------..((((((((((.(((.------------...)))...))))))))))...))))))))))).))))).... ( -44.70, z-score = -3.13, R) >droSim1.chr2L 8247872 107 + 22036055 UGGCCAAGGGUGUGCAAUUGCAUUGGCCAUAUCCACAUCCACAUCCAU------------------------GGACAUGGCCAUAGCC------------GUAGCUCUUUGGCCAUGGCAAUUGCGAUUGCAUUUUCUUUAUC .....(((((.(((((((((((.(((((((.((((............)------------------------))).)))))))..(((------------((.(((....))).)))))...))))))))))).))))).... ( -43.40, z-score = -3.44, R) >droSec1.super_3 3957641 107 + 7220098 UGGCCAAGGGUGUGCAAUUGCAUUGGCCAUAUCCACAUCCACAUCCAU------------------------GGACAUGGCCAUAGCC------------GUAGCUCUUUGGCCAUGGCAAUUGCGAUUGCAUUUUCUUUAUC .....(((((.(((((((((((.(((((((.((((............)------------------------))).)))))))..(((------------((.(((....))).)))))...))))))))))).))))).... ( -43.40, z-score = -3.44, R) >droEre2.scaffold_4929 9056873 143 + 26641161 UGACCAAGGGUGUGCAGUUGCUUUGGCCACAUCCACAUCCACAUCCAUAUCCAUGGCCAAAGCCACAUCCAUGGCCAUAGCCAUAGCCAUUGCCAUUGCCGUAGCUCUUGGGCCAUGGCAAUUGCGAUUGCAUUUUCUUUAUC .......(((((((.....((((((((((........................)))))))))))))))))(((((....))))).((.(((((.((((((((.(((....))).)))))))).))))).))............ ( -54.86, z-score = -3.70, R) >droVir3.scaffold_13049 16074427 108 - 25233164 CAGUCAGGGACAACAAGUUG----GGCCACGUCCACAUUCACGUCCAAGUCCA------------------CGUCCAUGUCCAUGUCCA----------CGUUGCACGCUGGUCAAAGCGAAGGC---UGCGUUUUGUUUGCC (((.((((((((.......(----(((...))))((((..((((........)------------------)))..))))...)))))(----------(((.((.((((......))))...))---.)))).))).))).. ( -27.00, z-score = 1.14, R) >consensus UGGCCAAGGGUGUGCAAUUGCAUUGGCCAUAUCCACAUCCACAUCCAU_UCCA___________________GGCCAUGGCCAUAGCC____________GUAGCUCUUUGGCCAUGGCAAUUGCGAUUGCAUUUUCUUUAUC .....(((((.((((((((((...((......)).......................................((((((((((.(((................)))...))))))))))....)))))))))).))))).... (-22.55 = -25.55 + 3.00)
| Location | 8,470,383 – 8,470,502 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 143 |
| Reading direction | reverse |
| Mean pairwise identity | 70.43 |
| Shannon entropy | 0.46490 |
| G+C content | 0.50200 |
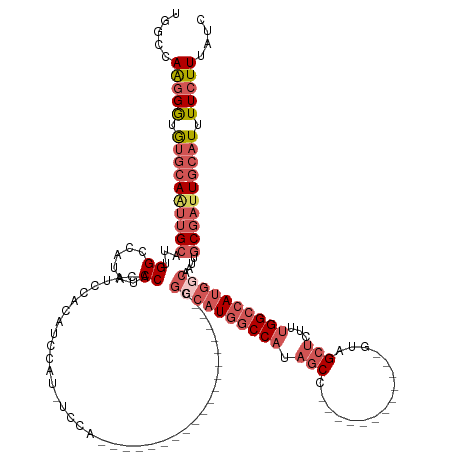
| Mean single sequence MFE | -44.14 |
| Consensus MFE | -20.78 |
| Energy contribution | -23.34 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.869453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 8470383 119 - 23011544 GAUAAAGAAAAUGCAAUCGCAAUUGCCAUGGCCAAAGAGCUAC------------GGCUAUGGCCAUGGCUA------------UGUCCAUGGAUGUGGAUGUGAAUGUGGAUAUGGCCAAUGCAAUUGCACACCCUUGGCCA ............(((.((((....((((((((((...(((...------------.))).))))))))))..------------.((((((....)))))))))).))).....((((((((((....))).....))))))) ( -50.90, z-score = -4.01, R) >droSim1.chr2L 8247872 107 - 22036055 GAUAAAGAAAAUGCAAUCGCAAUUGCCAUGGCCAAAGAGCUAC------------GGCUAUGGCCAUGUCC------------------------AUGGAUGUGGAUGUGGAUAUGGCCAAUGCAAUUGCACACCCUUGGCCA ............(((((....)))))..(((((((..(((...------------.))).(((((((((((------------------------(((........)))))))))))))).(((....))).....))))))) ( -42.50, z-score = -2.85, R) >droSec1.super_3 3957641 107 - 7220098 GAUAAAGAAAAUGCAAUCGCAAUUGCCAUGGCCAAAGAGCUAC------------GGCUAUGGCCAUGUCC------------------------AUGGAUGUGGAUGUGGAUAUGGCCAAUGCAAUUGCACACCCUUGGCCA ............(((((....)))))..(((((((..(((...------------.))).(((((((((((------------------------(((........)))))))))))))).(((....))).....))))))) ( -42.50, z-score = -2.85, R) >droEre2.scaffold_4929 9056873 143 - 26641161 GAUAAAGAAAAUGCAAUCGCAAUUGCCAUGGCCCAAGAGCUACGGCAAUGGCAAUGGCUAUGGCUAUGGCCAUGGAUGUGGCUUUGGCCAUGGAUAUGGAUGUGGAUGUGGAUGUGGCCAAAGCAACUGCACACCCUUGGUCA (((.(((....((((..((((....((((((((....(((((..((.((....)).))..)))))..)))))))).))))((((((((((((..((((........))))..))))))))))))...))))....))).))). ( -57.80, z-score = -2.42, R) >droVir3.scaffold_13049 16074427 108 + 25233164 GGCAAACAAAACGCA---GCCUUCGCUUUGACCAGCGUGCAACG----------UGGACAUGGACAUGGACG------------------UGGACUUGGACGUGAAUGUGGACGUGGCC----CAACUUGUUGUCCCUGACUG (((((.(((.(((..---((...((((......)))).))..))----------)((.((((..((((.(((------------------(........))))...))))..))))..)----)...))))))))........ ( -27.00, z-score = 1.82, R) >consensus GAUAAAGAAAAUGCAAUCGCAAUUGCCAUGGCCAAAGAGCUAC____________GGCUAUGGCCAUGGCC___________________UGGA_AUGGAUGUGGAUGUGGAUAUGGCCAAUGCAAUUGCACACCCUUGGCCA ...........((((.(((((.((((((((((((...((((..............)))).))))))))))...........................)).))))).))))....((((((((((....))).....))))))) (-20.78 = -23.34 + 2.56)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:26:09 2011