
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,815,100 – 19,815,202 |
| Length | 102 |
| Max. P | 0.751907 |

| Location | 19,815,100 – 19,815,202 |
|---|---|
| Length | 102 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 65.54 |
| Shannon entropy | 0.68850 |
| G+C content | 0.49860 |
| Mean single sequence MFE | -25.83 |
| Consensus MFE | -12.22 |
| Energy contribution | -11.97 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.07 |
| Mean z-score | -0.82 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.751907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 19815100 102 - 27905053 GCCACCUAGUGCCAGUGCUUAGAGCAGUUUCUCUGCAAGCAUUGAGACUGAGACUCAACUCGAU---UGGGACGGACCUGGAGGUGAAUCACACGCUGAAGAAAC------------ ..((((((((..((((((((...((((.....))))))))))))..)))(((......)))..(---..((.....))..))))))...................------------ ( -27.40, z-score = 0.23, R) >droSim1.chr3R 19639686 102 - 27517382 GCCACCUAGUGCCAGUGCUUAGAGCAGUUUCUGUGCAAGCAUUGAGACUGAGACUCGACUCGAU---UGGGACGAACCUGGAGGUGAAUCACACGCUGAAGGAAC------------ ........((.(((((.....((((((((((.(((....))).)))))))...)))......))---))).))...(((..(((((.....))).))..)))...------------ ( -28.30, z-score = 0.04, R) >droSec1.super_0 20156626 102 - 21120651 GCCACCUAGUGCCAGUGCUUAGAGCAGUUUCUGUGCAAGCAUUGAGACUGAGACUCGACUCGAU---UGGGACGAACCUGGAGGUGAAUCACACGCUGAAGGAAC------------ ........((.(((((.....((((((((((.(((....))).)))))))...)))......))---))).))...(((..(((((.....))).))..)))...------------ ( -28.30, z-score = 0.04, R) >droYak2.chr3R 2145409 97 + 28832112 GCCACCUAGUGCCAGUGCUUAGAGCAGUUUCUGUGCAAGCAUUGAGACUGAG-----ACUCGAU---UGGAAUGGACCUGGAGGUGAAUCACACGCUGAAGGAAC------------ .((..((((..(((...((..((((((((((.(((....))).)))))))..-----.)))...---.))..)))..))))(((((.....))).))...))...------------ ( -26.60, z-score = -0.09, R) >droEre2.scaffold_4820 2227982 90 + 10470090 GCCACCUAGUGCCAGUGCUUAGAGCAGUUUCUCUGCAAGCAUUGAGACUGA------------U---UGGAAUGGACUUGGGGGUGAAUCACACGCUGAAGGAAC------------ .(((..((((..((((((((...((((.....))))))))))))..)))).------------.---)))......(((.((.(((.....))).)).)))....------------ ( -29.70, z-score = -1.43, R) >droAna3.scaffold_13340 15953191 105 + 23697760 ACCACCUAGUGCCAGUGCUUGGAGCAGUUUCUGUGCAAGCAUUGAGACUGAAAUCGGGCGCGAUGGAAGGAAAGGAACCAGAGGAAACUUUUACAUUGACGGAUG------------ .((.(((....(((((((((((..(((((((.(((....))).)))))))...))))))))..))).))).........((((....)))).........))...------------ ( -33.00, z-score = -2.12, R) >dp4.chr2 14323008 81 - 30794189 ACCACCUAGUGCCAGUGCUUGGAGCAGUUUCUAUGCAAGCACUGAGACUG-----------------AUCUAGCAAUCGAGAAAG-AAGCACACGCUCC------------------ ........((((...((((.(((.(((((((..((....))..)))))))-----------------.))))))).((......)-).)))).......------------------ ( -23.20, z-score = -1.16, R) >droPer1.super_0 5713861 82 + 11822988 ACCACCUAGUGCCAGUGCUUGGAGCAGUUUCUAUGCAAGCACUGAGACUG-----------------AUCGAGCAAUCGAGAAAGAAAGCACACGCUCC------------------ ........(((((((((((((.(..........).)))))))))...((.-----------------.((((....))))...))...)))).......------------------ ( -26.40, z-score = -2.25, R) >droVir3.scaffold_13047 18550128 116 - 19223366 ACCACCUAGUGCCCGUGCUUGGAGCAGUUUCUGUGCACGCAUUGAGACUGAUAAAGACAAUUCAAA-AAAAAAAAAAACAACAGACGACCAAACACAGAACCGCACACAUAUGCAAA ........(((((.(((.((((..(((((((.(((....))).))))))).....((....))...-.....................)))).))).)....))))........... ( -22.20, z-score = -1.29, R) >droMoj3.scaffold_6540 7094879 82 - 34148556 GCCACCUAGUGCCCGUGCUUGGAGCAGUUUCUAUGCAUGCAUUGAGACUGAAC--------------AGCAAAUCACAUCGCACCGCACAUCACAA--------------------- ........((((..((((.((((.(((((((.(((....))).)))))))...--------------......)).))..)))).)))).......--------------------- ( -21.90, z-score = -1.08, R) >droGri2.scaffold_15074 1593603 102 + 7742996 GCCACCUAGUGCCCGUGCUUGGAGCAGUUUCUCUGCACGCACUGAGACUGAU---GAUAUUUAAGACAAUUAUUGCAAUUACACAUGACCAAACACAUAAGAA-AC----------- ..............(((.((((..(((((((..((....))..)))))))..---((((...........))))..............)))).))).......-..----------- ( -17.10, z-score = 0.11, R) >consensus GCCACCUAGUGCCAGUGCUUGGAGCAGUUUCUGUGCAAGCAUUGAGACUGAG______CUCGAU___AGGAAAGAACCUGGAGGUGAAUCACACGCUGAAGGAAC____________ .....((((.((....))))))..(((((((..((....))..)))))))................................................................... (-12.22 = -11.97 + -0.25)

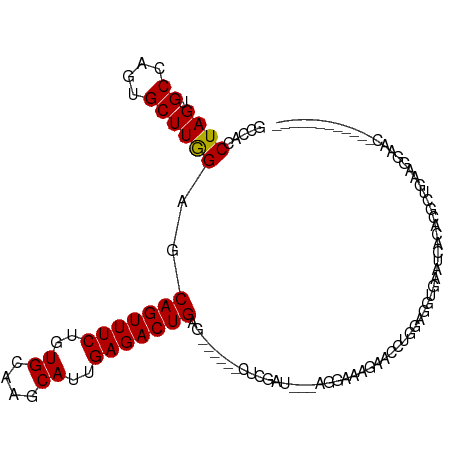
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:39:25 2011