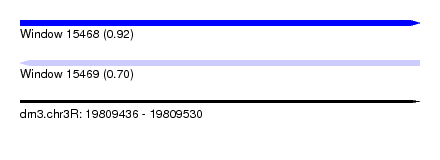
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,809,436 – 19,809,530 |
| Length | 94 |
| Max. P | 0.919567 |

| Location | 19,809,436 – 19,809,530 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 72.43 |
| Shannon entropy | 0.55760 |
| G+C content | 0.48309 |
| Mean single sequence MFE | -21.31 |
| Consensus MFE | -16.78 |
| Energy contribution | -15.79 |
| Covariance contribution | -0.98 |
| Combinations/Pair | 1.64 |
| Mean z-score | -0.77 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.919567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
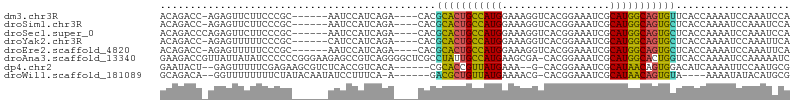
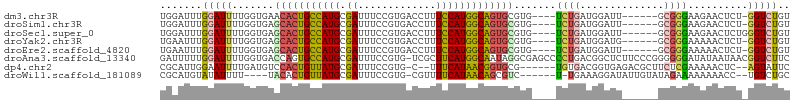
>dm3.chr3R 19809436 94 + 27905053 ACAGACC-AGAGUUCUUCCCGC------AAUCCAUCAGA----CACGCACUGCCAUGGAAAGGUCACGGAAAUCGCAUGGCAGUGUUCACCAAAAUCCAAAUCCA .......-..............------.........((----...(((((((((((((.............)).)))))))))))))................. ( -18.52, z-score = 0.04, R) >droSim1.chr3R 19633757 94 + 27517382 ACAGACC-AGAGUUCUUCCCGC------AAUCCAUCAGA----CACGCACUGCCAUGGAAAGGUCACGGAAAUCGCAUGGCAGUGCUCACCAAAAUCCAAAUCCA .......-..............------...........----...(((((((((((((.............)).)))))))))))................... ( -20.82, z-score = -0.68, R) >droSec1.super_0 20151018 95 + 21120651 ACAGACCCAGAGUUCUUCCCGC------AAUCCAUCAGA----CACGCACUGCCAUGGAAAGGUCACGGAAAUCGCAUGGCAGUGCUCACCAAAAUCCAAAUCCA ......................------...........----...(((((((((((((.............)).)))))))))))................... ( -20.82, z-score = -0.59, R) >droYak2.chr3R 2139683 94 - 28832112 ACAGACC-AGAGUUUUUCCCGC------CAUCCAUCAGA----CACGCACUGCCAUGGAAAGGUCACGGAAAUCGCAUGGCAGUGCUCACCAAAAUCCAAAUUCA .......-.((((((.......------..((.....))----...(((((((((((((.............)).)))))))))))............)))))). ( -22.32, z-score = -1.21, R) >droEre2.scaffold_4820 2222285 94 - 10470090 ACAGACC-AGAGUUUUUCCCGC------AAUCCAUCAGA----CACGCACUGCCAUGGAAAGGUCACGGAAAUCGCAUGGCAGUGCUCACCAAAAUCCAAAUUCA .......-.((((((.......------..((.....))----...(((((((((((((.............)).)))))))))))............)))))). ( -22.32, z-score = -1.33, R) >droAna3.scaffold_13340 15947530 104 - 23697760 GAAGACCGUUAUUAUAUCCCCCCGGGAAGAGCCGUCAGGGGCUCGCCUAUUGCCAUGAAGCGA-CACGGAAAUCGCAUGGCACUGGUCACCAAAAUCCAAAAAUC ...((((.........((((...)))).(((((......)))))......((((((...((((-........))))))))))..))))................. ( -30.90, z-score = -1.28, R) >dp4.chr2 14316080 94 + 30794189 GAAUACU--GAGUUUUUCGAGAAGCGUCUCACCGUCACA------CGCACCGUUAUGAAA--G-CACGGAAAUCGCAUAACAGUGGACAUCAAAAUUCCAAUGCG .......--(((((((..((((....))))...(((.((------(.....((((((..(--.-........)..)))))).))))))...)))))))....... ( -16.30, z-score = 0.76, R) >droWil1.scaffold_181089 11290358 91 - 12369635 GCAGACA--GGUUUUUUUUCUAUACAAUAUCCUUUCA-A------GACGCUGUUAUGAAAACG-CACGGAAAUCGCAUAACAGUGUA----AAAAUAUACAUGCG (((((.(--((...................))).)).-.------.(((((((((((....(.-...).......))))))))))).----..........))). ( -18.51, z-score = -1.87, R) >consensus ACAGACC_AGAGUUUUUCCCGC______AAUCCAUCAGA____CACGCACUGCCAUGGAAAGGUCACGGAAAUCGCAUGGCAGUGCUCACCAAAAUCCAAAUCCA ..............................................(((((((((((..................)))))))))))................... (-16.78 = -15.79 + -0.98)
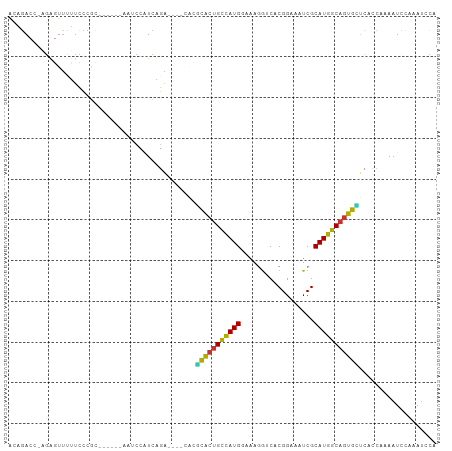
| Location | 19,809,436 – 19,809,530 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 72.43 |
| Shannon entropy | 0.55760 |
| G+C content | 0.48309 |
| Mean single sequence MFE | -27.92 |
| Consensus MFE | -16.76 |
| Energy contribution | -16.01 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.46 |
| Mean z-score | -0.80 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.704598 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
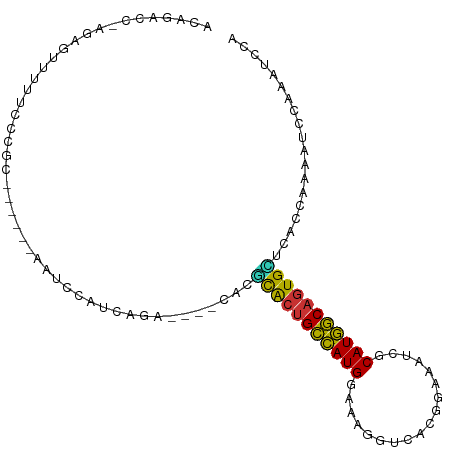
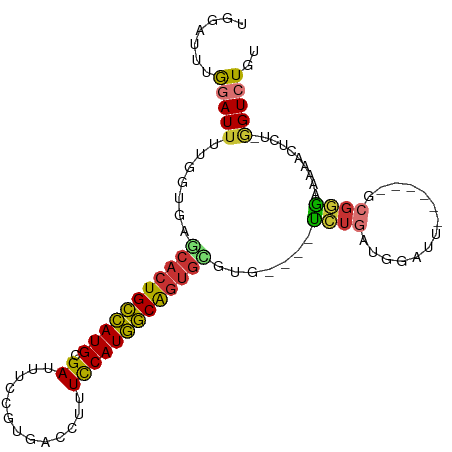
>dm3.chr3R 19809436 94 - 27905053 UGGAUUUGGAUUUUGGUGAACACUGCCAUGCGAUUUCCGUGACCUUUCCAUGGCAGUGCGUG----UCUGAUGGAUU------GCGGGAAGAACUCU-GGUCUGU .((((..(..((((......((((((((((.((.............))))))))))))((((----((.....)).)------)))...))))..).-.)))).. ( -25.62, z-score = 0.15, R) >droSim1.chr3R 19633757 94 - 27517382 UGGAUUUGGAUUUUGGUGAGCACUGCCAUGCGAUUUCCGUGACCUUUCCAUGGCAGUGCGUG----UCUGAUGGAUU------GCGGGAAGAACUCU-GGUCUGU .((((..(..((((.....(((((((((((.((.............)))))))))))))...----((((.......------.)))).))))..).-.)))).. ( -28.92, z-score = -0.55, R) >droSec1.super_0 20151018 95 - 21120651 UGGAUUUGGAUUUUGGUGAGCACUGCCAUGCGAUUUCCGUGACCUUUCCAUGGCAGUGCGUG----UCUGAUGGAUU------GCGGGAAGAACUCUGGGUCUGU .(((((..((((((.(..((((((((((((.((.............)))))))))))))...----((.....))).------.).))))....))..))))).. ( -32.62, z-score = -1.41, R) >droYak2.chr3R 2139683 94 + 28832112 UGAAUUUGGAUUUUGGUGAGCACUGCCAUGCGAUUUCCGUGACCUUUCCAUGGCAGUGCGUG----UCUGAUGGAUG------GCGGGAAAAACUCU-GGUCUGU .......(((((..(((..(((((((((((.((.............))))))))))))).((----((........)------)))......)))..-))))).. ( -29.72, z-score = -1.13, R) >droEre2.scaffold_4820 2222285 94 + 10470090 UGAAUUUGGAUUUUGGUGAGCACUGCCAUGCGAUUUCCGUGACCUUUCCAUGGCAGUGCGUG----UCUGAUGGAUU------GCGGGAAAAACUCU-GGUCUGU .......(((((..(((..(((((((((((.((.............)))))))))))))...----((((.......------.))))....)))..-))))).. ( -28.62, z-score = -1.07, R) >droAna3.scaffold_13340 15947530 104 + 23697760 GAUUUUUGGAUUUUGGUGACCAGUGCCAUGCGAUUUCCGUG-UCGCUUCAUGGCAAUAGGCGAGCCCCUGACGGCUCUUCCCGGGGGGAUAUAAUAACGGUCUUC .................((((..(((((((((((......)-))))...))))))......(((((......))))).((((...)))).........))))... ( -31.70, z-score = 0.10, R) >dp4.chr2 14316080 94 - 30794189 CGCAUUGGAAUUUUGAUGUCCACUGUUAUGCGAUUUCCGUG-C--UUUCAUAACGGUGCG------UGUGACGGUGAGACGCUUCUCGAAAAACUC--AGUAUUC ...(((((..((((((((((((((((((((((....((((.-.--.......))))..))------)))))))))).))))....))))))...))--))).... ( -30.20, z-score = -2.50, R) >droWil1.scaffold_181089 11290358 91 + 12369635 CGCAUGUAUAUUUU----UACACUGUUAUGCGAUUUCCGUG-CGUUUUCAUAACAGCGUC------U-UGAAAGGAUAUUGUAUAGAAAAAAAACC--UGUCUGC .(((((((......----))))...(((((((((.(((..(-((((........))))).------.-.....))).)))))))))..........--....))) ( -16.00, z-score = 0.05, R) >consensus UGGAUUUGGAUUUUGGUGAGCACUGCCAUGCGAUUUCCGUGACCUUUCCAUGGCAGUGCGUG____UCUGAUGGAUU______GCGGGAAAAACUCU_GGUCUGU ...................(((((((((((.((.............))))))))))))).............................................. (-16.76 = -16.01 + -0.75)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:39:25 2011