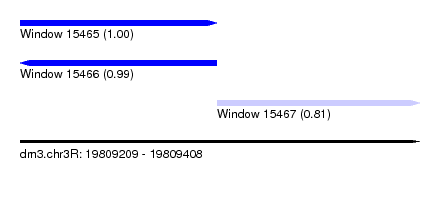
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,809,209 – 19,809,408 |
| Length | 199 |
| Max. P | 0.999966 |

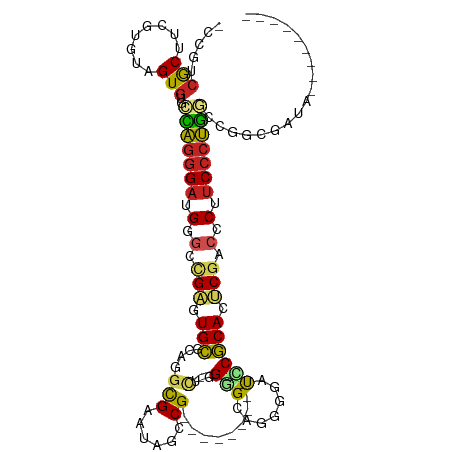
| Location | 19,809,209 – 19,809,307 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.28 |
| Shannon entropy | 0.61585 |
| G+C content | 0.64221 |
| Mean single sequence MFE | -50.36 |
| Consensus MFE | -27.08 |
| Energy contribution | -26.19 |
| Covariance contribution | -0.90 |
| Combinations/Pair | 1.72 |
| Mean z-score | -4.42 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 5.34 |
| SVM RNA-class probability | 0.999966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
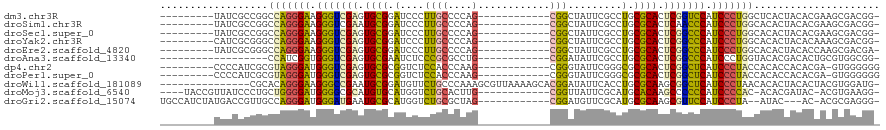
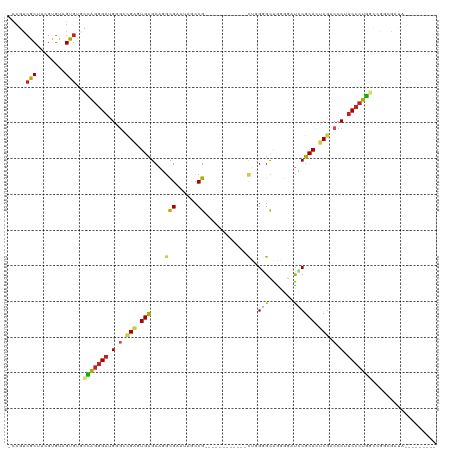
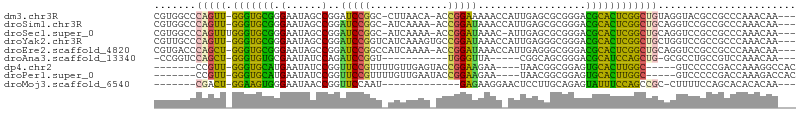
>dm3.chr3R 19809209 98 + 27905053 ---------UAUCGCCGGCCAGGGAAGGGUCGAGUGCGGAUCCCUUGCCCCAG------------CGGCUAUUCGCCUGCGCACUCGGUCCAUCCCUGGCUCACUACACGAAGCGACGG- ---------..((((..((((((((.((..(((((((((..........)).(------------((((.....))).)))))))))..)).))))))))((.......)).))))...- ( -49.40, z-score = -3.70, R) >droSim1.chr3R 19633530 98 + 27517382 ---------UAUCGCCGGCCAGGGAAGGGUCGAAUGCGGAUCCCUUGCCCCAG------------CGGCUAUUCGCCUGCGCACUCAACCCAUCCCUGGCACACUACACGAAGCGACGG- ---------..((((..((((((((.((((.((.(((((..........)).(------------((((.....))).))))).)).)))).))))))))...(.....)..))))...- ( -40.80, z-score = -2.68, R) >droSec1.super_0 20150791 98 + 21120651 ---------UAUCGCCGGCCAGGGAAGGGUCGAGUGCGGAUCCCUUGCCCCAG------------CGGCUAUUCGCCUGCGCACUCGGCCCAUCCCUGGCACACUACACGAAGCGACGG- ---------..((((..((((((((.(((((((((((((..........)).(------------((((.....))).))))))))))))).))))))))...(.....)..))))...- ( -52.10, z-score = -4.46, R) >droYak2.chr3R 2139454 98 - 28832112 ---------CAUCGCGGGCCAGGGAAGGGUCGAGUGCGGAUCCCUUGCCCCAG------------CGGCUAUUCGCCUGCGCACUCGGCCCAUCCCUGGCACACUACACAAAGCGACGG- ---------..((((..((((((((.(((((((((((((..........)).(------------((((.....))).))))))))))))).))))))))............))))...- ( -52.94, z-score = -4.76, R) >droEre2.scaffold_4820 2222057 98 - 10470090 ---------UAUCGCGGGCCAGGGAAGGGUCGAGUGCGGAUCCCUUGCCCCAG------------CGGCUAUUCGCCUGCGCACUCGGCCCAUCCCUGGCACACUACACCAAGCGACGA- ---------..((((..((((((((.(((((((((((((..........)).(------------((((.....))).))))))))))))).))))))))............))))...- ( -52.94, z-score = -4.68, R) >droAna3.scaffold_13340 15947329 89 - 23697760 ------------------CCAUCGGUGGGUCGAGUGCGAAUCUCCCGCGCCUG------------CGGAUAUUCGCCUGCGCACUCGGCCCAUCCCUGGUACACGACACUGCGUGGCGG- ------------------(((..((((((((((((((((((...((((....)------------)))..))))(....)))))))))))))))..)))..((((......))))....- ( -42.90, z-score = -2.74, R) >dp4.chr2 14315869 98 + 30794189 ---------CCCCAUCGCGUAGGGAUGGGUCGAGUGCGCGGUCUCCACCCAAG------------CGGGUAUUCGGGCGCGCACUCGGCUCAUCCCUACCACACCACACGA-GUGGGGGG ---------((((...(.((((((((((((((((((((((.((...((((...------------.))))....)).)))))))))))))))))))))))...((((....-)))))))) ( -65.80, z-score = -7.64, R) >droPer1.super_0 5706667 98 - 11822988 ---------CCCCAUCGCGUAGGGAUGGGUCGAGUGCGCGGUCUCCACCCAAG------------CGGGUAUUCGGGCGCGCACUCGGCUCAUCCCUACCACACCACACGA-GUGGGGGG ---------((((...(.((((((((((((((((((((((.((...((((...------------.))))....)).)))))))))))))))))))))))...((((....-)))))))) ( -65.80, z-score = -7.64, R) >droWil1.scaffold_181089 11290124 104 - 12369635 ---------------CGCACAGGGAAGGGCCGAAUGCGGAUGUUCUGCCCAAAGCGUUAAAAGCACGGAUAUUCACCUGCGCAAGCGGCUCAUCCCUAACACACUACACUACGUGGAUG- ---------------.....(((((.((((((..(((.((((((........))))))....(((.((.......))))))))..)))))).)))))......((((.....))))...- ( -34.80, z-score = -2.06, R) >droMoj3.scaffold_6540 7087306 101 + 34148556 ----UACCGUUAUCCCUGCUGGGGAUGGGGCGCAUGUGCAUGGUCUGCACUUG------------CGGUUAUUCGCAUGCACAAGCCCCCCAUCCCCAC-ACACGAUAC-ACGUGAAGG- ----..........(((..(((((((((((.((.(((((((((.((((....)------------))).....).)))))))).)).))))))))))).-.((((....-.)))).)))- ( -49.90, z-score = -4.75, R) >droGri2.scaffold_15074 1586716 101 - 7742996 UGCCAUCUAUGACCGUUGCCAGGGAUGGGAUGAAUGCGCAUGGUCUGCGCUAG------------CGGAUGUUCGCAUGCGCAAGCGUUCCAUCCCUA--AUAC---AC-ACGCGAGGG- ............((.((((.((((((((((((..((((((((((((((....)------------))))).....))))))))..)))))))))))).--....---..-..)))).))- ( -46.60, z-score = -3.48, R) >consensus _________UAUCGCCGGCCAGGGAAGGGUCGAGUGCGGAUCCCCUGCCCCAG____________CGGCUAUUCGCCUGCGCACUCGGCCCAUCCCUGGCACACUACACGAAGCGACGG_ ..................(((((((.(((((((.((((.........................................)))).))))))).)))))))..................... (-27.08 = -26.19 + -0.90)

| Location | 19,809,209 – 19,809,307 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.28 |
| Shannon entropy | 0.61585 |
| G+C content | 0.64221 |
| Mean single sequence MFE | -48.15 |
| Consensus MFE | -16.38 |
| Energy contribution | -16.14 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.57 |
| SVM RNA-class probability | 0.992781 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 19809209 98 - 27905053 -CCGUCGCUUCGUGUAGUGAGCCAGGGAUGGACCGAGUGCGCAGGCGAAUAGCCG------------CUGGGGCAAGGGAUCCGCACUCGACCCUUCCCUGGCCGGCGAUA--------- -((((((((......)))))((((((((.((..(((((((((.(((.....))))------------).(((........))))))))))..)).))))))))))).....--------- ( -49.50, z-score = -2.63, R) >droSim1.chr3R 19633530 98 - 27517382 -CCGUCGCUUCGUGUAGUGUGCCAGGGAUGGGUUGAGUGCGCAGGCGAAUAGCCG------------CUGGGGCAAGGGAUCCGCAUUCGACCCUUCCCUGGCCGGCGAUA--------- -.(((((((......)))(.((((((((.(((((((((((((.(((.....))))------------).(((........)))))))))))))).)))))))))))))...--------- ( -53.10, z-score = -3.82, R) >droSec1.super_0 20150791 98 - 21120651 -CCGUCGCUUCGUGUAGUGUGCCAGGGAUGGGCCGAGUGCGCAGGCGAAUAGCCG------------CUGGGGCAAGGGAUCCGCACUCGACCCUUCCCUGGCCGGCGAUA--------- -.(((((((......)))(.((((((((.(((.(((((((((.(((.....))))------------).(((........)))))))))).))).)))))))))))))...--------- ( -52.50, z-score = -2.83, R) >droYak2.chr3R 2139454 98 + 28832112 -CCGUCGCUUUGUGUAGUGUGCCAGGGAUGGGCCGAGUGCGCAGGCGAAUAGCCG------------CUGGGGCAAGGGAUCCGCACUCGACCCUUCCCUGGCCCGCGAUG--------- -.((((((.....)).(((.((((((((.(((.(((((((((.(((.....))))------------).(((........)))))))))).))).)))))))).)))))))--------- ( -53.20, z-score = -3.49, R) >droEre2.scaffold_4820 2222057 98 + 10470090 -UCGUCGCUUGGUGUAGUGUGCCAGGGAUGGGCCGAGUGCGCAGGCGAAUAGCCG------------CUGGGGCAAGGGAUCCGCACUCGACCCUUCCCUGGCCCGCGAUA--------- -..(((((.....)).(((.((((((((.(((.(((((((((.(((.....))))------------).(((........)))))))))).))).)))))))).)))))).--------- ( -52.00, z-score = -2.79, R) >droAna3.scaffold_13340 15947329 89 + 23697760 -CCGCCACGCAGUGUCGUGUACCAGGGAUGGGCCGAGUGCGCAGGCGAAUAUCCG------------CAGGCGCGGGAGAUUCGCACUCGACCCACCGAUGG------------------ -....((((......))))..(((.((.((((.((((((((....)((((.((((------------(....)))))..))))))))))).))))))..)))------------------ ( -37.60, z-score = -1.00, R) >dp4.chr2 14315869 98 - 30794189 CCCCCCAC-UCGUGUGGUGUGGUAGGGAUGAGCCGAGUGCGCGCCCGAAUACCCG------------CUUGGGUGGAGACCGCGCACUCGACCCAUCCCUACGCGAUGGGG--------- ((((((((-....))))((((.((((((((.(.((((((((((..(...(((((.------------...)))))..)..)))))))))).).))))))))))))..))))--------- ( -56.50, z-score = -5.25, R) >droPer1.super_0 5706667 98 + 11822988 CCCCCCAC-UCGUGUGGUGUGGUAGGGAUGAGCCGAGUGCGCGCCCGAAUACCCG------------CUUGGGUGGAGACCGCGCACUCGACCCAUCCCUACGCGAUGGGG--------- ((((((((-....))))((((.((((((((.(.((((((((((..(...(((((.------------...)))))..)..)))))))))).).))))))))))))..))))--------- ( -56.50, z-score = -5.25, R) >droWil1.scaffold_181089 11290124 104 + 12369635 -CAUCCACGUAGUGUAGUGUGUUAGGGAUGAGCCGCUUGCGCAGGUGAAUAUCCGUGCUUUUAACGCUUUGGGCAGAACAUCCGCAUUCGGCCCUUCCCUGUGCG--------------- -....(((........))).((((((((.(.((((..((((..((((..(..(((.((.......))..)))..)...))))))))..)))).).)))))).)).--------------- ( -32.10, z-score = -0.17, R) >droMoj3.scaffold_6540 7087306 101 - 34148556 -CCUUCACGU-GUAUCGUGU-GUGGGGAUGGGGGGCUUGUGCAUGCGAAUAACCG------------CAAGUGCAGACCAUGCACAUGCGCCCCAUCCCCAGCAGGGAUAACGGUA---- -......(((-...((.(((-.(((((((((((.((.((((((((.(......(.------------...)......))))))))).)).)))))))))))))).))...)))...---- ( -51.00, z-score = -3.78, R) >droGri2.scaffold_15074 1586716 101 + 7742996 -CCCUCGCGU-GU---GUAU--UAGGGAUGGAACGCUUGCGCAUGCGAACAUCCG------------CUAGCGCAGACCAUGCGCAUUCAUCCCAUCCCUGGCAACGGUCAUAGAUGGCA -......(((-.(---((..--(((((((((......((((((((.(....((.(------------(....)).)))))))))))......)))))))))))))))((((....)))). ( -35.60, z-score = -0.77, R) >consensus _CCGUCGCUUCGUGUAGUGUGCCAGGGAUGGGCCGAGUGCGCAGGCGAAUAGCCG____________CUGGGGCAGGGGAUCCGCACUCGACCCUUCCCUGGCCGGCGAUA_________ .....(((........)))..(((((((.(.(.(((.((((.........................................)))).))).).).))))))).................. (-16.38 = -16.14 + -0.24)
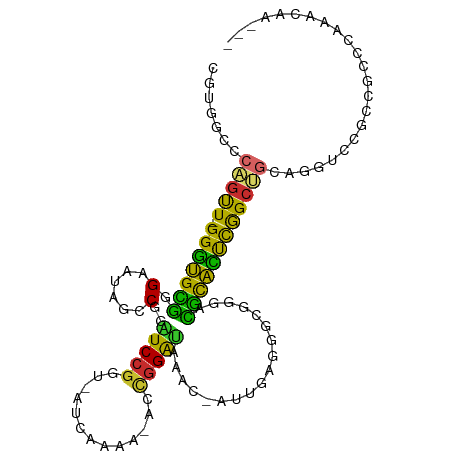
| Location | 19,809,307 – 19,809,408 |
|---|---|
| Length | 101 |
| Sequences | 9 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 64.15 |
| Shannon entropy | 0.72167 |
| G+C content | 0.59408 |
| Mean single sequence MFE | -34.96 |
| Consensus MFE | -16.93 |
| Energy contribution | -15.12 |
| Covariance contribution | -1.81 |
| Combinations/Pair | 1.94 |
| Mean z-score | -0.73 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.807507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
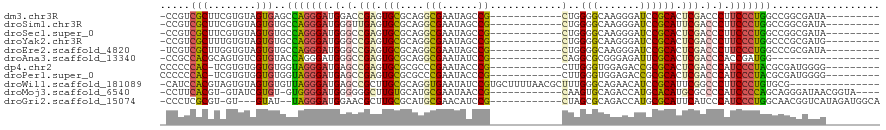
>dm3.chr3R 19809307 101 + 27905053 CGUGGCCCAGUU-GGGUGCGGGAAUAGCCGGAUCCGGC-CUUAACA-ACCGGAAAAACCAUUGAGCGCGGGACGCACUCGGCUGUAGGUACGCCGCCCAAACAA--- .(.(((.(((((-(((((((((.....))...(((.((-(((((..-...((.....)).))))).)).)))))))))))))))..((....))))))......--- ( -37.30, z-score = -0.14, R) >droSim1.chr3R 19633628 101 + 27517382 CGUGGCCCAGUU-GGGUGCGGGAAUAGCCGGAUCCGGC-AUCAAAA-ACCGGAUAAACCAUUGAGCGCGGGACGCACUCGGCUGCAGGUCCGCCGCCCAAACAA--- ..........((-(((((((((..(((((((((((((.-.......-.))))))...))...(((.(((...))).))))))))....)))).)))))))....--- ( -40.20, z-score = -0.79, R) >droSec1.super_0 20150889 101 + 21120651 CGUGGCCCAGUUUGGGUGCGGGAAUAGCCGGAUCCGGC-AUCAAAA-ACCGGAUAAAC-AUUGAGCGCGGGACGCACUCGGCUGCAGGUCCGCCGCCCAAACAA--- .........(((((((((((((..((((((.((((((.-.......-.))))))...)-...(((.(((...))).))))))))....)))).)))))))))..--- ( -43.90, z-score = -2.06, R) >droYak2.chr3R 2139552 103 - 28832112 CGUUGCCCAGUU-GGGUGCGGGAAUAGCCGGAUCCGGUCAUCAAAGUGCCGGAUAAACCAUUGAGGGCGGGACGCACUCGGCUGCUGGUCCGCCGCCCAAACAA--- ..........((-(((((((((..(((((((((((((.(((....)))))))))...))...(((.(((...))).))))))))....)))).)))))))....--- ( -42.40, z-score = -0.80, R) >droEre2.scaffold_4820 2222155 102 - 10470090 CGUGACCCAGCU-GGGUGCGGGAAUAGCCGGAUCCGGCCAUCAAAA-ACCGGAUAAACCAUUGAGGGCGGGACGCACUCGGCUGCAGGUCCGCCGCCCAAACAA--- ((.(((((((((-(((((((((.....))...(((.(((.((((..-...((.....)).)))).))).)))))))))))))))..))))))............--- ( -42.60, z-score = -1.50, R) >droAna3.scaffold_13340 15947418 85 - 23697760 -CCGGUCCAGCU-GGGUGUGCGAAUAUCCAGAUCCGGU-----------UGGGUUA-----CGGCAGCGGGACGCAUCCAGCUG-GCGCCUGCCGUCCAAACAA--- -.((((((((((-((((.((.(.....))).)))))))-----------)))....-----((.((((.(((....))).))))-.))...)))).........--- ( -35.20, z-score = -0.92, R) >dp4.chr2 14315967 90 + 30794189 -------CCGUU-GGGUGCAUGAAUAUCCGGUUCCGUUUUGUUGAGUACCGGAAGAA----UAACGGCGGAGUGCACUUGGC-----GUCCCCCGACCAAAGGCCAC -------((.((-((...........((((((.(((......)).).))))))....----...(((.((..(((.....))-----)..))))).)))).)).... ( -27.10, z-score = 0.16, R) >droPer1.super_0 5706765 90 - 11822988 -------CCGUU-GGGUGCAUGAAUAUCCGGUUCCGUUUUGUUGAAUACCGGAAGAA----UAACGGCGGAGUGCACUUGGC-----GUCCCCCGACCAAAGACCAC -------..(((-.(((.........((((((..((......))...))))))....----...(((.((..(((.....))-----)..))))))))...)))... ( -25.20, z-score = 0.13, R) >droMoj3.scaffold_6540 7087407 82 + 34148556 -------CGACU-GGAAGUGGGAAUAACCGGUUCCAAU-------------GAGAAGGAACUCCUUGCAGAGUAUUUCCAGCCGC-CUUUUCCAGCACACACAA--- -------((.((-((((((((((((.....)))))..(-------------(..((((....)))).))...))))))))).)).-..................--- ( -20.70, z-score = -0.63, R) >consensus CGUGGCCCAGUU_GGGUGCGGGAAUAGCCGGAUCCGGU_AUCAAAA_ACCGGAUAAAC_AUUGAGGGCGGGACGCACUCGGCUGCAGGUCCGCCGCCCAAACAA___ .......(((((.(((((((.(......)..(((((.............)))))..................))))))))))))....................... (-16.93 = -15.12 + -1.81)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:39:23 2011