
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,763,112 – 19,763,211 |
| Length | 99 |
| Max. P | 0.894611 |

| Location | 19,763,112 – 19,763,211 |
|---|---|
| Length | 99 |
| Sequences | 14 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 72.04 |
| Shannon entropy | 0.62810 |
| G+C content | 0.44555 |
| Mean single sequence MFE | -20.84 |
| Consensus MFE | -13.42 |
| Energy contribution | -12.56 |
| Covariance contribution | -0.86 |
| Combinations/Pair | 1.55 |
| Mean z-score | -0.68 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.894611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 19763112 99 - 27905053 CUGAACCACGUCUCGUACGGUCGCCUGACCUUCAAGUAUGAGCGCGAGUCCAACUACCAUCUGCUUGGUAAG--U----UUAUCACACUAUUAUUAUGCAAAACA----- .(((((..((((((((((((((....)))).....))))))).)))........(((((......))))).)--)----))).......................----- ( -21.40, z-score = -0.90, R) >droSim1.chr3R 19587202 99 - 27517382 CUGAACCACGUCUCGUACGGUCGCCUGACCUUCAAGUAUGAGCGCGAGUCCAACUACCAUCUGCUCGGUAAG--U----UUAUAUCACUAUUAUUAUCCAGAACA----- (((.....((.(((((((((((....)))).....))))))))).((....(((((((........))).))--)----)....))............)))....----- ( -21.10, z-score = -0.85, R) >droSec1.super_0 20104842 99 - 21120651 CUGAACCACGUCUCGUACGGUCGCCUGACCUUCAAGUAUGAGCGCGAAUCCAACUACCAUCUGCUCGGUAAG--U----UUAUAUCACUAUUAUUAUCCAAAACA----- .(((((..((((((((((((((....)))).....))))))).)))........((((........)))).)--)----))).......................----- ( -19.50, z-score = -1.38, R) >droYak2.chr3R 2092382 98 + 28832112 CUGAACCACGUCUCGUACGGUCGCCUGACCUUCAAGUACGAGCGCGAGUCCAACUACCAUCUGCUCGGUAAG-------UUAUGUCAUUAUUAUUAUACAGAACA----- (((.....((((((((((((((....)))).....))))))).))).....(((((((........))).))-------)).................)))....----- ( -23.10, z-score = -1.02, R) >droEre2.scaffold_4820 2175224 103 + 10470090 CUGAACCACGUCUCGUACGGUCGCCUGACCUUCAAGUAUGAGCGCGAGUCCAACUACCAUCUGCUCGGUAAG--UUAUAUUAUAUCACUAUUAUUAUCCAGAAUA----- (((.....((((((((((((((....)))).....))))))).))).....(((((((........))).))--))......................)))....----- ( -21.10, z-score = -0.69, R) >droAna3.scaffold_13340 15899721 98 + 23697760 CUCAACCACGUCUCGUACGGUCGCCUGACCUUCAAGUACGAGCGCGAGUCCAACUACCAUCUGCUCGGUGAG--UU---UCGAAUCGUACUCUAACUAAUAUG------- .........(.(((((((((((....)))).....))))))))((((.((.(((((((........))).))--))---..)).))))...............------- ( -23.50, z-score = -0.93, R) >droPer1.super_0 5646870 103 + 11822988 CUCAACCAUGUCUCGUACGGUCGCCUGACCUUCAAGUACGAGCGCGAGUCCAACUAUCAUUUGCUCGGUAAGUCUCUCGUUCUAUAUUUCGAUCCAUCUAUUG------- .........(.(((((((((((....)))).....))))))))(((((....((((((........))).)))..))))).......................------- ( -20.20, z-score = -0.77, R) >dp4.chr2 14264020 103 - 30794189 CUCAACCAUGUCUCGUACGGUCGCCUGACCUUCAAGUACGAACGCGAGUCCAACUAUCAUUUGCUCGGUAAGUCUCUCGUUCUAUAUUUUGAUCUAUCUAUUG------- .((((.........((((((((....)))).....))))(((((.(((.(..(((...........)))..).))).)))))......))))...........------- ( -19.00, z-score = -0.77, R) >droWil1.scaffold_181089 11237417 108 + 12369635 CUCAACCAUGUCUCAUACGGUCGCUUGACCUUCAAGUAUGAACGUGAAUCCAACUACCAUUUGCUCGGUAAG--UUGAAUAAGAAAAUCUUUCAAAUGGUUCUUAGCACU ......(((((.((((((((((....)))).....)))))))))))....((((((((........))).))--)))..(((((..(((........))))))))..... ( -24.50, z-score = -2.10, R) >droVir3.scaffold_13047 10951305 91 + 19223366 CUCAAUCAUGUCUCAUACGGUCGUCUGACCUUCAAGUAUGAGCGCGAGUCCAACUAUCAUCUGCUCGGUAAG-UU--AGUCGAGU-----UCCAUAGAG----------- .....((.((.(((((((((((....)))).....))))))))).))......((((.....((((((....-..--..))))))-----...))))..----------- ( -21.30, z-score = -0.90, R) >droMoj3.scaffold_6540 27919368 96 - 34148556 CUGAACCAUGUCUCAUACGGUCGCCUGACCUUCAAAUAUGAGCGCGAAUCCAACUAUCAUCUGCUCGGUAAG-UU--GUUAGGGUCUUUAUUUAUAUAG----------- ..((.((.(((((((((.((((....))))......)))))).)))....((((((((........))).))-))--)...)).)).............----------- ( -16.80, z-score = 0.48, R) >droGri2.scaffold_15074 1337998 95 - 7742996 CUGAACCAUGUCUCAUACGGUCGCCUGACUUUCAAGUAUGAGCGCGAGUCCAACUACCAUUUGCUCGGUAAG-UUUGAGUUUAAGAGUUAUUCACA-------------- .........(((((((((((((....)))).....))))))).))((((..((((.......((((((....-.)))))).....))))))))...-------------- ( -19.70, z-score = 0.12, R) >anoGam1.chr2R 56856150 96 - 62725911 CUGAACCACGUCUCGUACGGCCGUCUGACGUUCAAGUACGAGCGAGAGUCGAACUACCAUCUGCUCGGUGAG--UU---UUCCAGCAUUGCAUGCAUACAG--------- (((.(((.((.((((((((..((.....))..)..))))))))).((((.((.......)).)))))))...--..---.....((((...))))...)))--------- ( -25.10, z-score = 0.18, R) >apiMel3.Group14 283246 106 - 8318479 UUGAAUCAUGUGUCGUAUGGUCGUCUAACUUUUAAAUAUGAAAAAGAAUCAAAUUAUCACUUAUUAGGUAUAUACAUAAUUCAUAUAGUUGGCAUCCAUUUGAAUA---- ............(((.((((..(.((((((.....(((((((............((((........)))).........))))))))))))))..)))).)))...---- ( -15.50, z-score = 0.07, R) >consensus CUGAACCACGUCUCGUACGGUCGCCUGACCUUCAAGUAUGAGCGCGAGUCCAACUACCAUCUGCUCGGUAAG__U____UUAUAUCAUUAUUAAUAUACAGAA_______ ....(((.(((.((((((((((....)))).....))))))))).((((.............)))))))......................................... (-13.42 = -12.56 + -0.86)

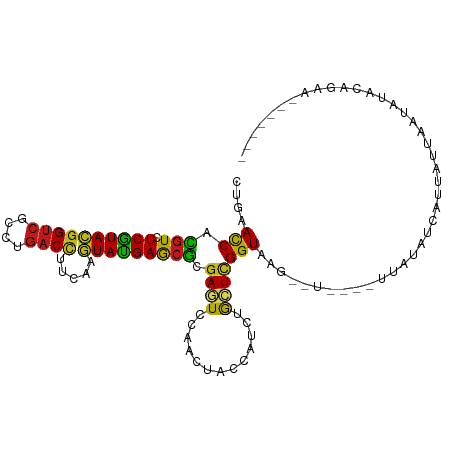

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:39:17 2011