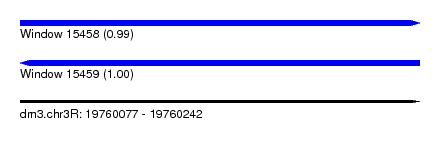
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,760,077 – 19,760,242 |
| Length | 165 |
| Max. P | 0.999729 |

| Location | 19,760,077 – 19,760,242 |
|---|---|
| Length | 165 |
| Sequences | 5 |
| Columns | 184 |
| Reading direction | forward |
| Mean pairwise identity | 67.30 |
| Shannon entropy | 0.56082 |
| G+C content | 0.44153 |
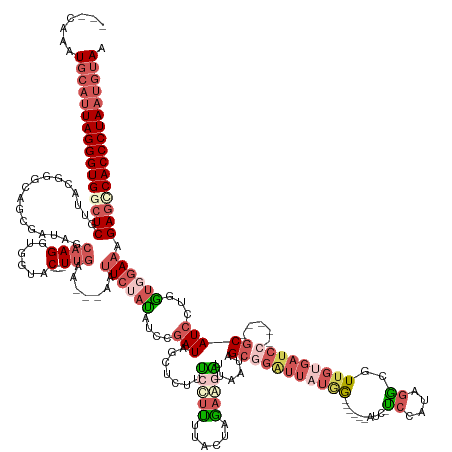
| Mean single sequence MFE | -48.58 |
| Consensus MFE | -20.98 |
| Energy contribution | -19.94 |
| Covariance contribution | -1.04 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.54 |
| SVM RNA-class probability | 0.992475 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
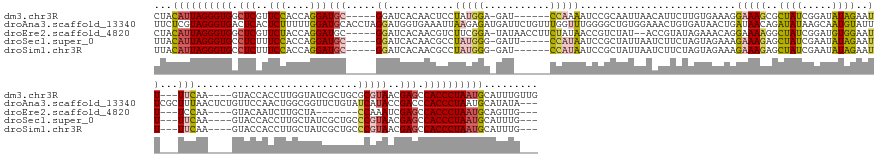
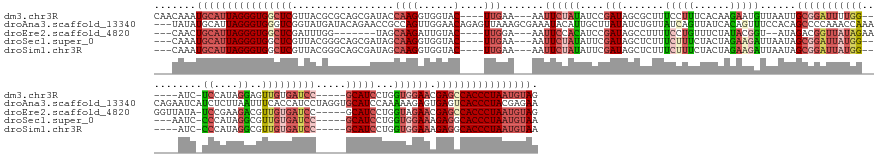
>dm3.chr3R 19760077 165 + 27905053 CUACAUUAGGGUGGCUCGUUCCACCAGGAUGC-----GGAUCACAACUCCUAUGGA-GAU------CCAAAAUCCGCAAUUAACAUUCUUGUGAAAGGAAAGCGCUAUCGGAUAUAGAAUU---UUCAA----GUACCACCUUGGUAUCGCUGCGCGUAACGAGCCACCCUAAUGCAUUUGUUG ...(((((((((((((((((.(.((....(((-----((((.....((((...)))-)..------.....)))))))....(((....)))....))...((((...((......((...---.))..----(((((.....)))))))..))))).)))))))))))))))))......... ( -59.80, z-score = -5.04, R) >droAna3.scaffold_13340 15896564 181 - 23697760 UUCUCGUAGGGUGACUCACUCUUUUUGGAUGCACCUAGGAUGGUGAAAUUAAGAGAUGAUUCUGUUUGGUUUGGGGCUGUGGAAACUGUGAUAACUGAUAACAGAUAUAAGCAAUGUAUUUCGCUUUAACUCUGUUCCAACUGGCGGUUCUGUAUCAUACCGACCCACCCUAAUGCAUAUA--- ......(((((((..((((((((..(.....((((......))))..)..))))).))).((.((.((((.((((((((((....).............((((((...((((..........))))....)))))).......))))))))).)))).)).))..))))))).........--- ( -46.40, z-score = -0.33, R) >droEre2.scaffold_4820 2172201 159 - 10470090 CUACAUUAGGGUGGCUCGUUCUACCAGGAUGC-----GGAUCACAACGUCUUCGGA-UAUAACCUUCUAUAACCGUCUAU--ACCGUAUAGAAACAGGAAAAGGCUAUCGGAUGUGGAAUU---UCCAA----GUACAAUCUUGCUA-------CCAAAUCGAGCCACCCUAAUGCAGUUG--- ((.(((((((((((((((((((((..(((((.-----.(....)..)))))((.((-((...((((......((.(((((--(...))))))....))..)))).)))).)).))))))..---..(((----(......))))...-------......))))))))))))))).))...--- ( -50.40, z-score = -3.62, R) >droSec1.super_0 20101805 163 + 21120651 UUACAUUAGGGUGCCUCUUUCCACCAGGAUGC-----GGAUCACAACGCCUAUGGG-GAUU-----CCAUAAUCCGCUAUUAAUCUUCUAGUAGAAAGAAAGAGCUAUCGAAUAUAGAAUU---UUCAA----GUACCACCUUGCUAUCGCUGCCCGUAACGAGCCACCCUAAUGCAUUUG--- ...((((((((((.(((..(((....)))(((-----((............(((((-((((-----....))))).))))..........((((...(((((..((((.....))))..))---))).(----(((......))))....)))))))))..))).))))))))))......--- ( -43.50, z-score = -2.13, R) >droSim1.chr3R 19584140 162 + 27517382 UUACAUUAGGGUGCCUCUUUCCACCAGGAUGC-----GGAUCACAACGCCUAUGGG-GAU------CCAUAAUCCGCUAUUAAUCUUCUAGUAGAAAGAAAGAGCUAUCGAAUAUAGAAUU---UUCAA----GUACCACCUUGCUAUCGCUGCCCGUAACGAGCCACCCUAAUGCAUUUG--- ...((((((((((.(((..(((....)))(((-----((............(((((-(((------.....)))).))))..........((((...(((((..((((.....))))..))---))).(----(((......))))....)))))))))..))).))))))))))......--- ( -42.80, z-score = -2.00, R) >consensus UUACAUUAGGGUGCCUCGUUCCACCAGGAUGC_____GGAUCACAACGCCUAUGGA_GAU______CCAUAAUCCGCUAUUAACCUUCUAGUAAAAGGAAAGAGCUAUCGGAUAUAGAAUU___UUCAA____GUACCACCUUGCUAUCGCUGCCCGUAACGAGCCACCCUAAUGCAUUUG___ ...((((((((((.(((..(((....)))........((....................................((((.........))))............((((.....))))...................)).......................))).))))))))))......... (-20.98 = -19.94 + -1.04)
| Location | 19,760,077 – 19,760,242 |
|---|---|
| Length | 165 |
| Sequences | 5 |
| Columns | 184 |
| Reading direction | reverse |
| Mean pairwise identity | 67.30 |
| Shannon entropy | 0.56082 |
| G+C content | 0.44153 |
| Mean single sequence MFE | -57.11 |
| Consensus MFE | -24.59 |
| Energy contribution | -27.19 |
| Covariance contribution | 2.60 |
| Combinations/Pair | 1.41 |
| Mean z-score | -3.88 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.27 |
| SVM RNA-class probability | 0.999729 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
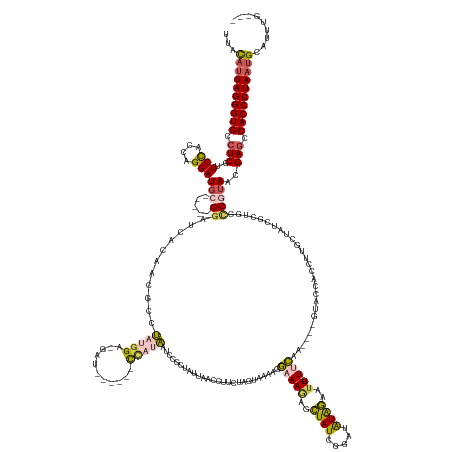
>dm3.chr3R 19760077 165 - 27905053 CAACAAAUGCAUUAGGGUGGCUCGUUACGCGCAGCGAUACCAAGGUGGUAC----UUGAA---AAUUCUAUAUCCGAUAGCGCUUUCCUUUCACAAGAAUGUUAAUUGCGGAUUUUGG------AUC-UCCAUAGGAGUUGUGAUCC-----GCAUCCUGGUGGAACGAGCCACCCUAAUGUAG .......(((((((((((((((((((.(((.(((.(((......((((...----..(((---(...((((.....))))...))))...)))).............(((((((....------..(-(((...))))....)))))-----))))))))))).))))))))))))))))))). ( -64.70, z-score = -5.41, R) >droAna3.scaffold_13340 15896564 181 + 23697760 ---UAUAUGCAUUAGGGUGGGUCGGUAUGAUACAGAACCGCCAGUUGGAACAGAGUUAAAGCGAAAUACAUUGCUUAUAUCUGUUAUCAGUUAUCACAGUUUCCACAGCCCCAAACCAAACAGAAUCAUCUCUUAAUUUCACCAUCCUAGGUGCAUCCAAAAAGAGUGAGUCACCCUACGAGAA ---.........(((((((((.((((..........))))))..((((((((((....((((((......))))))...))))))....((....))..................))))......(((.(((((.....((((......))))........))))))))..)))))))...... ( -40.82, z-score = -0.89, R) >droEre2.scaffold_4820 2172201 159 + 10470090 ---CAACUGCAUUAGGGUGGCUCGAUUUGG-------UAGCAAGAUUGUAC----UUGGA---AAUUCCACAUCCGAUAGCCUUUUCCUGUUUCUAUACGGU--AUAGACGGUUAUAGAAGGUUAUA-UCCGAAGACGUUGUGAUCC-----GCAUCCUGGUAGAACGAGCCACCCUAAUGUAG ---...((((((((((((((((((.((((.-------(((((((......)----)))..---.....((((..((((((((((((.((((((.(((...))--).))))))....)))))))))).-((....)))).))))....-----.....))).)))).)))))))))))))))))) ( -57.30, z-score = -4.42, R) >droSec1.super_0 20101805 163 - 21120651 ---CAAAUGCAUUAGGGUGGCUCGUUACGGGCAGCGAUAGCAAGGUGGUAC----UUGAA---AAUUCUAUAUUCGAUAGCUCUUUCUUUCUACUAGAAGAUUAAUAGCGGAUUAUGG-----AAUC-CCCAUAGGCGUUGUGAUCC-----GCAUCCUGGUGGAAAGAGGCACCCUAAUGUAA ---....((((((((((((.(((.....((((..(((...((((......)----)))..---..........)))...))))....((((((((((..(((.....((((((((..(-----...(-(.....))..)..))))))-----)))))))))))))))))).)))))))))))). ( -61.26, z-score = -4.33, R) >droSim1.chr3R 19584140 162 - 27517382 ---CAAAUGCAUUAGGGUGGCUCGUUACGGGCAGCGAUAGCAAGGUGGUAC----UUGAA---AAUUCUAUAUUCGAUAGCUCUUUCUUUCUACUAGAAGAUUAAUAGCGGAUUAUGG------AUC-CCCAUAGGCGUUGUGAUCC-----GCAUCCUGGUGGAAAGAGGCACCCUAAUGUAA ---....((((((((((((.(((.....((((..(((...((((......)----)))..---..........)))...))))....((((((((((..(((.....((((((((..(------..(-(.....))..)..))))))-----)))))))))))))))))).)))))))))))). ( -61.46, z-score = -4.35, R) >consensus ___CAAAUGCAUUAGGGUGGCUCGUUACGGGCAGCGAUAGCAAGGUGGUAC____UUGAA___AAUUCUAUAUCCGAUAGCUCUUUCCUUUUACUAGAAGAUUAAUAGCGGAUUAUGG______AUC_UCCAUAGGCGUUGUGAUCC_____GCAUCCUGGUGGAAAGAGCCACCCUAAUGUAA .......(((((((((((((((((((.............................................(((((.........(((((......))))).......)))))................(((((((...(((..........))).))).))))))))))))))))))))))). (-24.59 = -27.19 + 2.60)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:39:16 2011