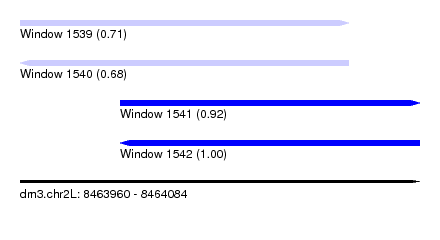
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,463,960 – 8,464,084 |
| Length | 124 |
| Max. P | 0.996178 |

| Location | 8,463,960 – 8,464,062 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 86.62 |
| Shannon entropy | 0.22092 |
| G+C content | 0.50520 |
| Mean single sequence MFE | -31.04 |
| Consensus MFE | -23.50 |
| Energy contribution | -24.02 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.714770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 8463960 102 + 23011544 UCCGCUUUAAUCAAGAGUCCGCGGGCAAUUGAAAAAGCUCUCGCUUCGCGUGUUCACGGCGUGUACCUACAUAGGUACGCAUCAAUUAGGUACCUGCCGCCC-------- ..(((.....((((..(((....)))..))))..((((....)))).))).......(((((((((((....))))))))........(((....)))))).-------- ( -31.90, z-score = -1.71, R) >droSim1.chr2L 8241480 102 + 22036055 UCCGCUUUAAUCAAGAGUCCGCGGGCAAUUGAAAAAGCUCUCGCUUCGCGUGUUCACGGCGUGUACCUACAUAGGUACGCCUCAAUUAGGUACCUGCCGCCC-------- ..(((.....((((..(((....)))..))))..((((....)))).)))......(((((.(((((((...(((....)))....))))))).)))))...-------- ( -32.80, z-score = -1.91, R) >droSec1.super_3 3951309 102 + 7220098 UCCGCUUUAAUCAAGAGUCCGCGGGCAAUUGAAAAAGCUCUCGCUUCGCGUGUUCACGGCGUGUACCUACAUAGGUACGCCUCAAUUAGGUACCUGCCGCCC-------- ..(((.....((((..(((....)))..))))..((((....)))).)))......(((((.(((((((...(((....)))....))))))).)))))...-------- ( -32.80, z-score = -1.91, R) >droYak2.chr2L 11093174 110 + 22324452 UCCGCUUUAAUCAACAGUCCGCGGGCAAUUGAAAAAGCUCUCGCUUUUCAUGUUGACCGCAUGUACCUACAAAGGUACGCACCAAUUAGGUACAACGCGCGUUCUAAGCA ...((((........((..(((((.((((((((((.((....)))))))).)))).))((.((((((((....(((....)))...))))))))..)))))..)))))). ( -33.20, z-score = -2.85, R) >droEre2.scaffold_4929 9050304 110 + 26641161 UCCGCUUUAAUCAACAGUCCGCGGGCAAUUGAAAAAGCUUUCGCUUCGCAUGUUCACCGCAUGUACCUACAUAGGUACGCAUCAAUUAAGUACAAUGCGCGUUCUAAACA ..(((...............((((..(((((...((((....))))..)).)))..))))..((((((....))))))((((............)))))))......... ( -24.50, z-score = -0.88, R) >consensus UCCGCUUUAAUCAAGAGUCCGCGGGCAAUUGAAAAAGCUCUCGCUUCGCGUGUUCACGGCGUGUACCUACAUAGGUACGCAUCAAUUAGGUACCUGCCGCCC________ ...(((((..((((..(((....)))..)))).)))))...................(((((((((((....))))))))........((......)))))......... (-23.50 = -24.02 + 0.52)

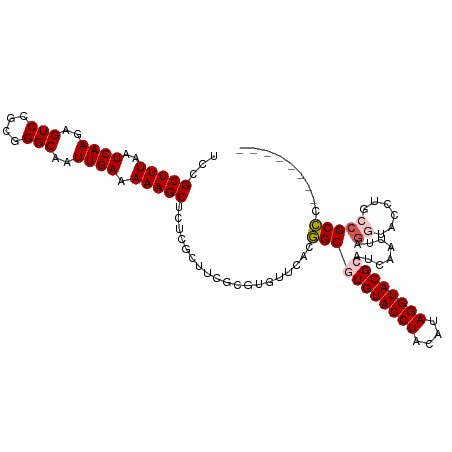
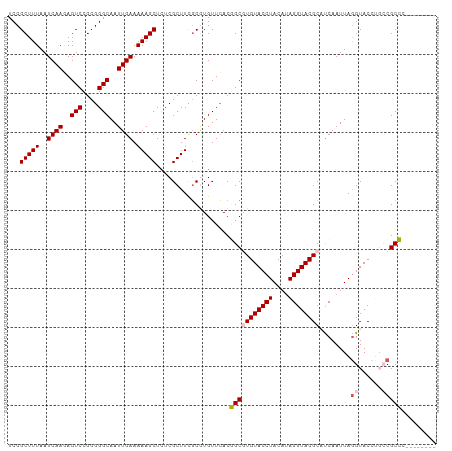
| Location | 8,463,960 – 8,464,062 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 86.62 |
| Shannon entropy | 0.22092 |
| G+C content | 0.50520 |
| Mean single sequence MFE | -35.84 |
| Consensus MFE | -27.38 |
| Energy contribution | -26.34 |
| Covariance contribution | -1.04 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.684392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 8463960 102 - 23011544 --------GGGCGGCAGGUACCUAAUUGAUGCGUACCUAUGUAGGUACACGCCGUGAACACGCGAAGCGAGAGCUUUUUCAAUUGCCCGCGGACUCUUGAUUAAAGCGGA --------(((((((..(((((((...(........)....)))))))..)))(((....)))(((((....))))).......))))((...............))... ( -31.46, z-score = -0.48, R) >droSim1.chr2L 8241480 102 - 22036055 --------GGGCGGCAGGUACCUAAUUGAGGCGUACCUAUGUAGGUACACGCCGUGAACACGCGAAGCGAGAGCUUUUUCAAUUGCCCGCGGACUCUUGAUUAAAGCGGA --------(((((((..(((((((....(((....)))...)))))))..)))(((....)))(((((....))))).......))))((...............))... ( -36.16, z-score = -1.66, R) >droSec1.super_3 3951309 102 - 7220098 --------GGGCGGCAGGUACCUAAUUGAGGCGUACCUAUGUAGGUACACGCCGUGAACACGCGAAGCGAGAGCUUUUUCAAUUGCCCGCGGACUCUUGAUUAAAGCGGA --------(((((((..(((((((....(((....)))...)))))))..)))(((....)))(((((....))))).......))))((...............))... ( -36.16, z-score = -1.66, R) >droYak2.chr2L 11093174 110 - 22324452 UGCUUAGAACGCGCGUUGUACCUAAUUGGUGCGUACCUUUGUAGGUACAUGCGGUCAACAUGAAAAGCGAGAGCUUUUUCAAUUGCCCGCGGACUGUUGAUUAAAGCGGA .........(((((((.(((((.....)))))((((((....))))))))))((.(((...(((((((....)))))))...))).)))))..(((((......))))). ( -40.70, z-score = -3.22, R) >droEre2.scaffold_4929 9050304 110 - 26641161 UGUUUAGAACGCGCAUUGUACUUAAUUGAUGCGUACCUAUGUAGGUACAUGCGGUGAACAUGCGAAGCGAAAGCUUUUUCAAUUGCCCGCGGACUGUUGAUUAAAGCGGA ((((((...(((((((((........))))))((((((....))))))..))).))))))((((((((....))))).(((((..((...))...))))).....))).. ( -34.70, z-score = -2.02, R) >consensus ________GGGCGGCAGGUACCUAAUUGAUGCGUACCUAUGUAGGUACACGCCGUGAACACGCGAAGCGAGAGCUUUUUCAAUUGCCCGCGGACUCUUGAUUAAAGCGGA .........((((....(((((((...(........)....))))))).))))..........(((((....))))).........((((...............)))). (-27.38 = -26.34 + -1.04)

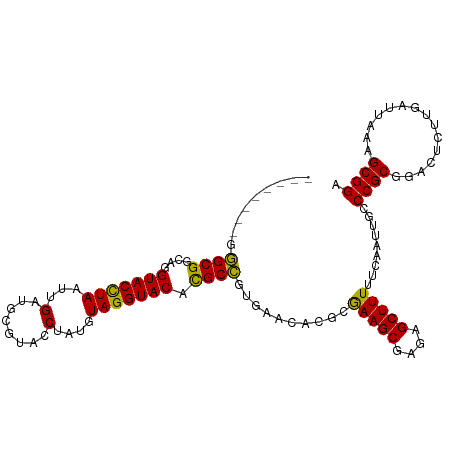
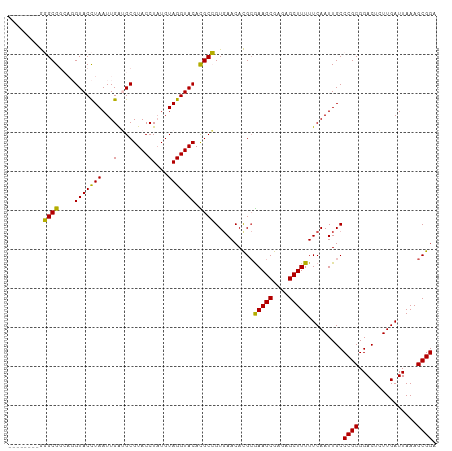
| Location | 8,463,991 – 8,464,084 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 72.96 |
| Shannon entropy | 0.39391 |
| G+C content | 0.47311 |
| Mean single sequence MFE | -26.66 |
| Consensus MFE | -17.02 |
| Energy contribution | -17.65 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.919086 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
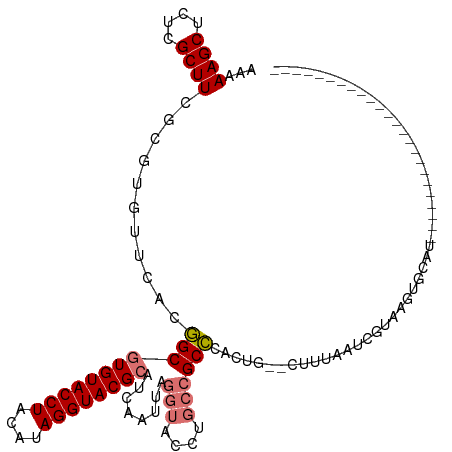
>dm3.chr2L 8463991 93 + 23011544 AAAAAGCUCUCGCUUCGCGUGUUCACGGCGUGUACCUACAUAGGUACGCAUCAAUUAGGUACCUGCCGCCCACUG--CUUUAAUCGUAAGUGCAU------------------------ ...((((....)))).(((((....((((((((((((....)))))))).......((....))))))..))).(--(((.......))))))..------------------------ ( -26.10, z-score = -1.41, R) >droSim1.chr2L 8241511 93 + 22036055 AAAAAGCUCUCGCUUCGCGUGUUCACGGCGUGUACCUACAUAGGUACGCCUCAAUUAGGUACCUGCCGCCCACUG--CUUUAAUCGUAAGUGCAU------------------------ ...((((....)))).(((((....(((((.(((((((...(((....)))....))))))).)))))..))).(--(((.......))))))..------------------------ ( -27.90, z-score = -2.08, R) >droSec1.super_3 3951340 85 + 7220098 AAAAAGCUCUCGCUUCGCGUGUUCACGGCGUGUACCUACAUAGGUACGCCUCAAUUAGGUACCUGCCGCCCACUG--AUUUAUUCGU-------------------------------- ...((((....))))...(((....(((((.(((((((...(((....)))....))))))).)))))..)))..--..........-------------------------------- ( -24.60, z-score = -2.11, R) >droEre2.scaffold_4929 9050335 119 + 26641161 AAAAAGCUUUCGCUUCGCAUGUUCACCGCAUGUACCUACAUAGGUACGCAUCAAUUAAGUACAAUGCGCGUUCUAAACAAUAACUUUAAUUUCCUGAGCUGCUUUAAUGGUACAUGCAA ...((((....)))).((((((..(((((..((((((....))))))((((............))))))((((......................)))).........))))))))).. ( -28.05, z-score = -2.05, R) >consensus AAAAAGCUCUCGCUUCGCGUGUUCACGGCGUGUACCUACAUAGGUACGCAUCAAUUAGGUACCUGCCGCCCACUG__CUUUAAUCGUAAGUGCAU________________________ ...((((....))))...........(((((((((((....))))))))........(((....))))))................................................. (-17.02 = -17.65 + 0.63)

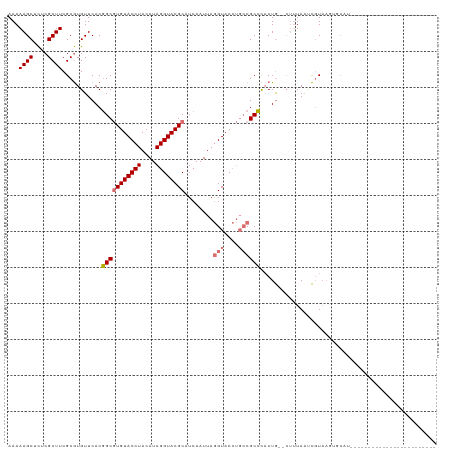
| Location | 8,463,991 – 8,464,084 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 72.96 |
| Shannon entropy | 0.39391 |
| G+C content | 0.47311 |
| Mean single sequence MFE | -32.98 |
| Consensus MFE | -24.75 |
| Energy contribution | -24.37 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.89 |
| SVM RNA-class probability | 0.996178 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
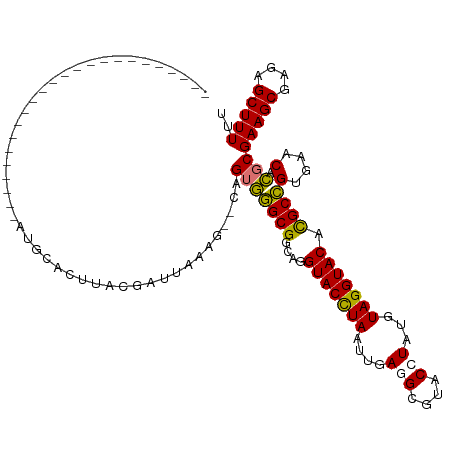
>dm3.chr2L 8463991 93 - 23011544 ------------------------AUGCACUUACGAUUAAAG--CAGUGGGCGGCAGGUACCUAAUUGAUGCGUACCUAUGUAGGUACACGCCGUGAACACGCGAAGCGAGAGCUUUUU ------------------------.................(--(.(((.(((((..(((((((...(........)....)))))))..)))))...)))))(((((....))))).. ( -29.80, z-score = -1.61, R) >droSim1.chr2L 8241511 93 - 22036055 ------------------------AUGCACUUACGAUUAAAG--CAGUGGGCGGCAGGUACCUAAUUGAGGCGUACCUAUGUAGGUACACGCCGUGAACACGCGAAGCGAGAGCUUUUU ------------------------.................(--(.(((.(((((..(((((((....(((....)))...)))))))..)))))...)))))(((((....))))).. ( -34.50, z-score = -3.06, R) >droSec1.super_3 3951340 85 - 7220098 --------------------------------ACGAAUAAAU--CAGUGGGCGGCAGGUACCUAAUUGAGGCGUACCUAUGUAGGUACACGCCGUGAACACGCGAAGCGAGAGCUUUUU --------------------------------..........--..(((.(((((..(((((((....(((....)))...)))))))..)))))...)))..(((((....))))).. ( -32.50, z-score = -3.56, R) >droEre2.scaffold_4929 9050335 119 - 26641161 UUGCAUGUACCAUUAAAGCAGCUCAGGAAAUUAAAGUUAUUGUUUAGAACGCGCAUUGUACUUAAUUGAUGCGUACCUAUGUAGGUACAUGCGGUGAACAUGCGAAGCGAAAGCUUUUU ..(((((((((....((((((((...........)))...))))).....((((((((........))))))((((((....))))))..)))))..))))))(((((....))))).. ( -35.10, z-score = -2.35, R) >consensus ________________________AUGCACUUACGAUUAAAG__CAGUGGGCGGCAGGUACCUAAUUGAGGCGUACCUAUGUAGGUACACGCCGUGAACACGCGAAGCGAGAGCUUUUU ..............................................(((((((....(((((((....(((....)))...))))))).))))(....).)))(((((....))))).. (-24.75 = -24.37 + -0.38)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:26:07 2011