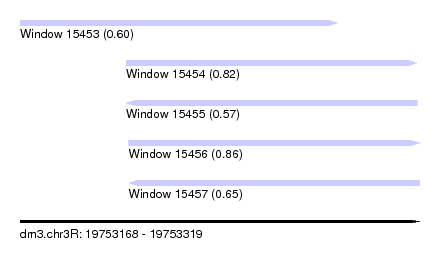
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,753,168 – 19,753,319 |
| Length | 151 |
| Max. P | 0.856773 |

| Location | 19,753,168 – 19,753,288 |
|---|---|
| Length | 120 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.85 |
| Shannon entropy | 0.55399 |
| G+C content | 0.48556 |
| Mean single sequence MFE | -36.63 |
| Consensus MFE | -15.51 |
| Energy contribution | -15.25 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.57 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.596412 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
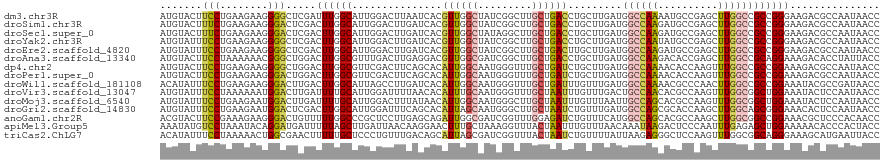
>dm3.chr3R 19753168 120 + 27905053 AUGUACUUCCUGAAGAAGGGGCUCGAUUUGGCAUUGGACUUAAUCACGUUGGCUAUCGGCUUGCUGACCUGCUUGAUGGCCAAAAUGCCGAGCUUGGCCGCCGGGAAGACGCCAAUAACC ..((.(((((((.....(.(((.((((((((((((.((.....))...((((((((((((..........)).))))))))))))))))))).)))))).)))))))))).......... ( -42.90, z-score = -2.15, R) >droSim1.chr3R 19577247 120 + 27517382 AUGUACUUUCUGAAGAAGGGACUCGACUUGGCAUUGGACUUGAUCACGUUGGCUAUCGGCUUGCUGACCUGCUUGAUGGCCAAGAUGCCGAGCUUGGCCGCCGGGAAGACGCCAAUAACC ..((.(((((((.....((...(((((((((((((.((.....))...((((((((((((..........)).))))))))))))))))))).))))))..))))))))).......... ( -37.30, z-score = -0.96, R) >droSec1.super_0 20094910 120 + 21120651 AUGUACUUUCUGAAGAAGGGACUCGACUUGGCAUUGGACUUGAUCACGUUGGCUAUAGGCUUGCUGACCUGCUUGAUGGCCAAGAUGCCGAGCUUGGCCGCCGGGAAGACGCCAAUAACC ..((.(((((....))))).))((..((((((.......(..(.((.((..((.........))..)).)).)..).(((((((........)))))))))))))..))........... ( -35.40, z-score = -0.59, R) >droYak2.chr3R 20646902 120 + 28832112 AUGUAUUUCCUGAAGAAGGGGCUCGACUUGGCAUUGGACUUGAUCACGUUGGCUAUCGGCUUGCUGACCUGCUUGAUGGCCAAUAUGCCGAGCUUGGCCGCCGGGAAGACGCCAAUAACC ..((.(((((((.....(.(((.(((((((((((..((.....))..(((((((((((((..........)).))))))))))))))))))).)))))).)))))))))).......... ( -42.40, z-score = -1.87, R) >droEre2.scaffold_4820 2165282 120 - 10470090 AUGUAUUUCCUGAAGAAGGGGCUCGACUUGGCAUUGGACUUGAUCACGUUGGCUAUCGGCUUGCUGACUUGCUUGAUGGCCAAGAUGCCGAGCUUGGCCGCCGGGAAGACGCCAAUAACC ..((.(((((((.....(.(((.((((((((((((.((.....))...((((((((((((..(....)..)).))))))))))))))))))).)))))).)))))))))).......... ( -43.40, z-score = -2.06, R) >droAna3.scaffold_13340 15890359 120 - 23697760 AUGUACUUCCUAAAAAACGGGCUGGACUUGGCGUUUGACUUGAGGACGUUGGCGAUCGGCUUGCUGACCUGCUUGAUGGCCAAGACACCGAGCUUGGCCGCAGGAAAGACACCUAUUACC .(((..(((((......(((((.((..(..((((((.......))))))..)...((((....)))))).)))))..(((((((........)))))))..)))))..)))......... ( -38.80, z-score = -1.97, R) >dp4.chr2 16898450 120 - 30794189 AUGUACUUCCUGAAGAAGGGGCUGGACUUGGCGUUCGACUUCAGCACAUUGGCAAUGGGUUUGCUGAUCUGCUUGAUGGCCAAAACACCAAGUUUGGCCGCCGGAAAGACGCCAAUAACC ..((..((((.......(((((..(((((((.(((...(.((((((.((..((((.....))))..)).))).))).).....))).)))))))..))).))))))..)).......... ( -41.81, z-score = -1.86, R) >droPer1.super_0 6549390 120 + 11822988 AUGUACUUCCUGAAGAAGGGACUGGACUUGGCGUUCGACUUCAGCACAUUGGCAAUGGGUUUGCUGAUCUGCUUGAUGGCCAAAACACCAAGUUUGGCCGCCGGAAAGACGCCAAUAACC ......(((((.....)))))..((..((((((((.....((((((.((..((((.....))))..)).))).))).(((((((........)))))))........))))))))...)) ( -39.00, z-score = -1.60, R) >droWil1.scaffold_181108 3977957 120 - 4707319 ACAUAUUUCCUGAAGAAGGGACUUGACUUGGCAUUAGCCUUGAUCACAUUGGCAAUGGGUUUGCUGAUUUGUUUGAUGGCCAAAACGCCCAACUUGGCCGCCGGAAAUACGCCGAUAACC ...(((((((.....(((........)))(((....((((..(.((.((..((((.....))))..)).)).)..).)))......(((......))).))))))))))........... ( -31.30, z-score = -0.50, R) >droVir3.scaffold_13047 13988024 120 + 19223366 AUGUAUUUCCUAAAAAAUGGACUUGAUUUUGCAUUGGAUUUUAACACAUUUGCAAUGGGUUUGCUAAUUUGUUUGACUGCCAACACGCCAAGUUUGGCGGCUGGAAAUACUCCAAUAACC ..((((((((...(((((.......)))))((.((((.....((((.(((.((((.....)))).))).))))......))))..(((((....))))))).)))))))).......... ( -29.30, z-score = -1.51, R) >droMoj3.scaffold_6540 8050462 120 - 34148556 AUGUAUUUCCUGAAGAAUGGACUUGAUUUUGCAUUGGACUUUAUAACAUUGGCAAUGGGCUUGCUAAUUUGUUUAAUUGCCAGCACGCCAAGUUUGGCGGCUGGAAAUACUCCAAUAACC .......(((........)))...........((((((....((((.((((((((.....))))))))))))...(((.(((((.(((((....)))))))))).)))..)))))).... ( -32.20, z-score = -1.55, R) >droGri2.scaffold_14830 299443 120 - 6267026 AUGUAUUUCCUGAAGAAUGGACUCGACUUGGCAUUGGAUUUCAGCACAUUAGCAAUGGGCUUGCUAAUCUGUUUGAUGGCCAGCGCACCAAGCUUGGCAGCGGGAAACACUCCAAUAACC ..((.(((((((.....((...(((((((((..((((...((((((.((((((((.....)))))))).)).))))...))))....))))).)))))).))))))).)).......... ( -31.70, z-score = 0.02, R) >anoGam1.chr2R 4193282 120 + 62725911 ACGUACUUCCGAAAGAAGGGACUGUUUUUGGCCCGCUCCUUGAGCAGAUUGGCGAUCGGUUUGGAGAUCUGUUUCAUGGCCAGCACGCCAAGCUUGGCGGCCGGAAACGCUCCCACAACC ..((.....(....)..((((.(((((((((((.(((((.((((((((((..(((.....)))..)))))).)))).))..)))..((((....))))))))))))))).)))))).... ( -47.60, z-score = -2.34, R) >apiMel3.Group5 9184219 120 + 13386189 AAAUAUGUCCUAAAUACAGGAUGAUUUUUAGCUUGAUUAACAAGGAACUUUGCUAAAGGUUUACUAAUUUGUUUAACAAAUAAGACUCCCAAUUUGAGAGCUGGAAAAACACCCACUACC ......(((((......)))))..((((((((((..........(((((((....))))))).((.(((((.....))))).)).............))))))))))............. ( -22.20, z-score = -1.34, R) >triCas2.ChLG7 12183152 120 + 17478683 ACAUAUUUCCUAAAAACUGGCGAACUUUUUGCUCCCUGUUUGACAGCAUUAGCGAUCGGUUUACUAAUCUGUUUUAUUAAGAGGGCUCCAAGUUUGGCGGCAGGGAAAGCAUGAAUUACC .(((.((((((.....(((.(((((((...((((.((....(((((.(((((..(.....)..))))))))))......)).))))...))))))).)))..))))))..)))....... ( -34.10, z-score = -1.97, R) >consensus AUGUAUUUCCUGAAGAAGGGACUCGACUUGGCAUUGGACUUGAUCACAUUGGCAAUCGGCUUGCUGAUCUGCUUGAUGGCCAAGACGCCAAGCUUGGCCGCCGGAAAGACGCCAAUAACC .......(((........))).....((((((...............((((((.........)))))).........((((((..........))))))))))))............... (-15.51 = -15.25 + -0.26)

| Location | 19,753,208 – 19,753,318 |
|---|---|
| Length | 110 |
| Sequences | 14 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 67.37 |
| Shannon entropy | 0.70124 |
| G+C content | 0.45692 |
| Mean single sequence MFE | -28.27 |
| Consensus MFE | -12.40 |
| Energy contribution | -11.59 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.65 |
| Mean z-score | -1.03 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.822208 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
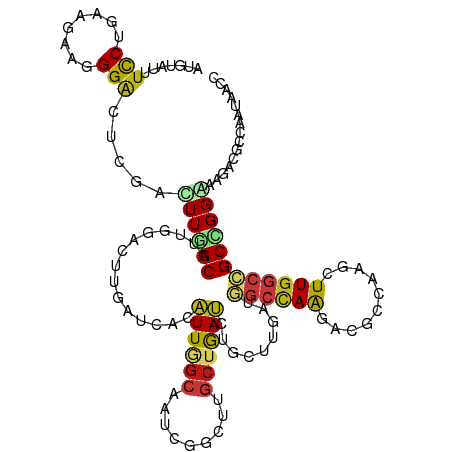
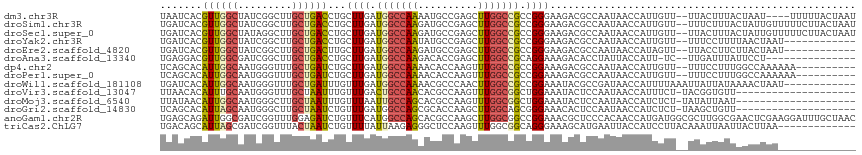
>dm3.chr3R 19753208 110 + 27905053 UAAUCACGUUGGCUAUCGGCUUGCUGACCUGCUUGAUGGCCAAAAUGCCGAGCUUGGCCGCCGGGAAGACGCCAAUAACCAUUGUU--UUACUUUACUAAU----UUUUUACUAAU .......((((((..((((....)))).((.((((.(((((((..........))))))).)))).))..))))))..........--.............----........... ( -25.20, z-score = -0.38, R) >droSim1.chr3R 19577287 114 + 27517382 UGAUCACGUUGGCUAUCGGCUUGCUGACCUGCUUGAUGGCCAAGAUGCCGAGCUUGGCCGCCGGGAAGACGCCAAUAACCAUUGUU--UUUCUUUACUAUUGUUUUUCUUACUAAU .((..(((.(((.(((.(((...((..((.((.....(((((((........))))))))).))..))..))).))).))).))).--..))........................ ( -29.70, z-score = -0.99, R) >droSec1.super_0 20094950 114 + 21120651 UGAUCACGUUGGCUAUAGGCUUGCUGACCUGCUUGAUGGCCAAGAUGCCGAGCUUGGCCGCCGGGAAGACGCCAAUAACCAUUGUU--UUACUUUACUAUUGUUUUUCUUACUAAU .....(((.(((.(((.(((...((..((.((.....(((((((........))))))))).))..))..))).))).))).))).--............................ ( -29.00, z-score = -0.91, R) >droYak2.chr3R 20646942 102 + 28832112 UGAUCACGUUGGCUAUCGGCUUGCUGACCUGCUUGAUGGCCAAUAUGCCGAGCUUGGCCGCCGGGAAGACGCCAAUAACCAUUGUU--UUUCCUUUUAACUAAU------------ .......(((((((((((((..........)).)))))))))))..((((....))))....(((((((((...........))))--)))))...........------------ ( -28.00, z-score = -0.98, R) >droEre2.scaffold_4820 2165322 102 - 10470090 UGAUCACGUUGGCUAUCGGCUUGCUGACUUGCUUGAUGGCCAAGAUGCCGAGCUUGGCCGCCGGGAAGACGCCAAUAACCAUAGUU--UUACCUUCUUACUAAU------------ .......((((((..((((....))))(((.((((.((((((((........)))))))).)))))))..)))))).....((((.--..........))))..------------ ( -30.20, z-score = -1.44, R) >droAna3.scaffold_13340 15890399 98 - 23697760 UGAGGACGUUGGCGAUCGGCUUGCUGACCUGCUUGAUGGCCAAGACACCGAGCUUGGCCGCAGGAAAGACACCUAUUACCAUU-UC--UUGAUUUAUUCCU--------------- .(((((...(((..((.((..((((..(((((.....(((((((........))))))))))))..)).)))).))..))).)-))--))...........--------------- ( -29.90, z-score = -2.07, R) >dp4.chr2 16898490 104 - 30794189 UCAGCACAUUGGCAAUGGGUUUGCUGAUCUGCUUGAUGGCCAAAACACCAAGUUUGGCCGCCGGAAAGACGCCAAUAACCAUUGUU--UUUCCUUUGGCCAAAAAA---------- ((((((.((..((((.....))))..)).))).)))(((........)))..((((((((..(((((((((...........))))--)))))..))))))))...---------- ( -35.30, z-score = -2.31, R) >droPer1.super_0 6549430 104 + 11822988 UCAGCACAUUGGCAAUGGGUUUGCUGAUCUGCUUGAUGGCCAAAACACCAAGUUUGGCCGCCGGAAAGACGCCAAUAACCAUUGUU--UUUCCUUUGGCCAAAAAA---------- ((((((.((..((((.....))))..)).))).)))(((........)))..((((((((..(((((((((...........))))--)))))..))))))))...---------- ( -35.30, z-score = -2.31, R) >droWil1.scaffold_181108 3977997 104 - 4707319 UGAUCACAUUGGCAAUGGGUUUGCUGAUUUGUUUGAUGGCCAAAACGCCCAACUUGGCCGCCGGAAAUACGCCGAUAACCAUUUUAAAUUAUUAUAAAACUAAU------------ .......(((((((((.((....)).)))..((((.(((((((..........))))))).)))).....))))))............................------------ ( -20.40, z-score = 0.27, R) >droVir3.scaffold_13047 13988064 95 + 19223366 UUAACACAUUUGCAAUGGGUUUGCUAAUUUGUUUGACUGCCAACACGCCAAGUUUGGCGGCUGGAAAUACUCCAAUAACCAUUUCU-UACGGUGUU-------------------- ..(((((....(.(((((..................(((((((.((.....))))))))).((((.....))))....))))).).-....)))))-------------------- ( -22.60, z-score = -0.71, R) >droMoj3.scaffold_6540 8050502 95 - 34148556 UUAUAACAUUGGCAAUGGGCUUGCUAAUUUGUUUAAUUGCCAGCACGCCAAGUUUGGCGGCUGGAAAUACUCCAAUAACCAUCUCU-UAUAUUAAU-------------------- .(((((.((((((((.....)))))))).......(((.(((((.(((((....)))))))))).))).................)-)))).....-------------------- ( -24.20, z-score = -1.63, R) >droGri2.scaffold_14830 299483 95 - 6267026 UCAGCACAUUAGCAAUGGGCUUGCUAAUCUGUUUGAUGGCCAGCGCACCAAGCUUGGCAGCGGGAAACACUCCAAUAACCAUCUCU-UAAGCUGUU-------------------- .((((..((((((((.....))))))))......(((((((((.((.....)))))))....(((.....)))......))))...-...))))..-------------------- ( -26.80, z-score = -0.47, R) >anoGam1.chr2R 4193322 116 + 62725911 UGAGCAGAUUGGCGAUCGGUUUGGAGAUCUGUUUCAUGGCCAGCACGCCAAGCUUGGCGGCCGGAAACGCUCCCACAACCAUGAUGGCGCUUGGCGAACUCGAAGGAUUUGCUAAC ..((((((((..(((..((((..((((....))))..))))....((((((((.(((.(((.(....)))).)))...((.....)).))))))))...)))...))))))))... ( -41.50, z-score = -1.27, R) >triCas2.ChLG7 12183192 103 + 17478683 UGACAGCAUUAGCGAUCGGUUUACUAAUCUGUUUUAUUAAGAGGGCUCCAAGUUUGGCGGCAGGGAAAGCAUGAAUUACCAUCCUUACAAAUUAAUUACUUAA------------- .(((((.(((((..(.....)..)))))))))).......((((((((((....))).))).((..............))..)))).................------------- ( -17.64, z-score = 0.71, R) >consensus UGAUCACAUUGGCAAUCGGCUUGCUGAUCUGCUUGAUGGCCAAGACGCCAAGCUUGGCCGCCGGAAAGACGCCAAUAACCAUUGUU__UUACUUUUUUACUAA_____________ .......((((((.........))))))...((((.(((((((..........))))))).))))................................................... (-12.40 = -11.59 + -0.80)
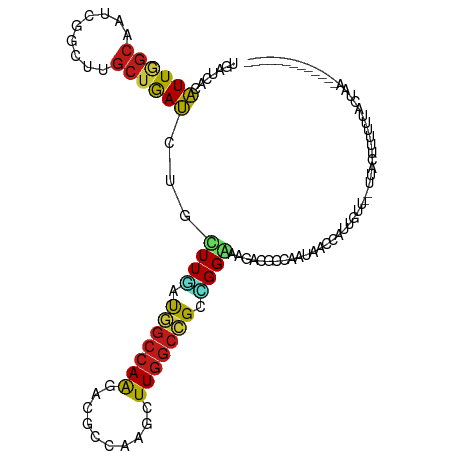
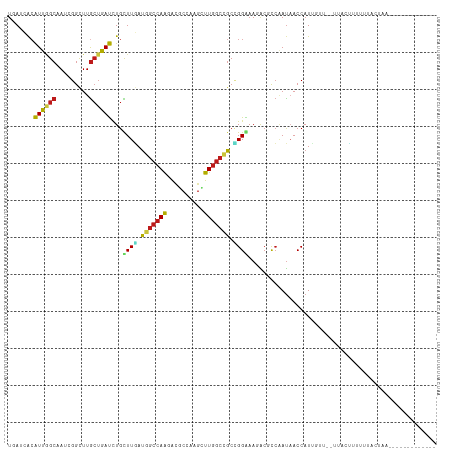
| Location | 19,753,208 – 19,753,318 |
|---|---|
| Length | 110 |
| Sequences | 14 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 67.37 |
| Shannon entropy | 0.70124 |
| G+C content | 0.45692 |
| Mean single sequence MFE | -29.92 |
| Consensus MFE | -9.77 |
| Energy contribution | -9.96 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.571015 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 19753208 110 - 27905053 AUUAGUAAAAA----AUUAGUAAAGUAA--AACAAUGGUUAUUGGCGUCUUCCCGGCGGCCAAGCUCGGCAUUUUGGCCAUCAAGCAGGUCAGCAAGCCGAUAGCCAACGUGAUUA ((((((.....----)))))).......--..((.(((((((((((..((..((.(((((((((........))))))).....)).))..))...)))))))))))...)).... ( -31.90, z-score = -1.93, R) >droSim1.chr3R 19577287 114 - 27517382 AUUAGUAAGAAAAACAAUAGUAAAGAAA--AACAAUGGUUAUUGGCGUCUUCCCGGCGGCCAAGCUCGGCAUCUUGGCCAUCAAGCAGGUCAGCAAGCCGAUAGCCAACGUGAUCA ........................((..--.((..(((((((((((..((..((.(((((((((........))))))).....)).))..))...)))))))))))..))..)). ( -34.10, z-score = -2.46, R) >droSec1.super_0 20094950 114 - 21120651 AUUAGUAAGAAAAACAAUAGUAAAGUAA--AACAAUGGUUAUUGGCGUCUUCCCGGCGGCCAAGCUCGGCAUCUUGGCCAUCAAGCAGGUCAGCAAGCCUAUAGCCAACGUGAUCA ........((...((....((.......--.))..(((((((.(((..((..((.(((((((((........))))))).....)).))..))...))).)))))))..))..)). ( -29.70, z-score = -1.26, R) >droYak2.chr3R 20646942 102 - 28832112 ------------AUUAGUUAAAAGGAAA--AACAAUGGUUAUUGGCGUCUUCCCGGCGGCCAAGCUCGGCAUAUUGGCCAUCAAGCAGGUCAGCAAGCCGAUAGCCAACGUGAUCA ------------............((..--.((..((((((((((((((.....)))((((..(((.(((......)))....))).)))).....)))))))))))..))..)). ( -31.80, z-score = -1.65, R) >droEre2.scaffold_4820 2165322 102 + 10470090 ------------AUUAGUAAGAAGGUAA--AACUAUGGUUAUUGGCGUCUUCCCGGCGGCCAAGCUCGGCAUCUUGGCCAUCAAGCAAGUCAGCAAGCCGAUAGCCAACGUGAUCA ------------...........(((..--.((..((((((((((((((.....)))(((((((........))))))).....((......))..)))))))))))..)).))). ( -34.80, z-score = -2.54, R) >droAna3.scaffold_13340 15890399 98 + 23697760 ---------------AGGAAUAAAUCAA--GA-AAUGGUAAUAGGUGUCUUUCCUGCGGCCAAGCUCGGUGUCUUGGCCAUCAAGCAGGUCAGCAAGCCGAUCGCCAACGUCCUCA ---------------((((.........--..-..(((..((.(((..((..(((((((((((((.....).))))))).....)))))..))...))).))..)))...)))).. ( -30.89, z-score = -1.87, R) >dp4.chr2 16898490 104 + 30794189 ----------UUUUUUGGCCAAAGGAAA--AACAAUGGUUAUUGGCGUCUUUCCGGCGGCCAAACUUGGUGUUUUGGCCAUCAAGCAGAUCAGCAAACCCAUUGCCAAUGUGCUGA ----------.....(((((((((.(..--..((..((((..(((((((.....))).)))))))))).).)))))))))...((((.((..((((.....))))..)).)))).. ( -31.60, z-score = -0.63, R) >droPer1.super_0 6549430 104 - 11822988 ----------UUUUUUGGCCAAAGGAAA--AACAAUGGUUAUUGGCGUCUUUCCGGCGGCCAAACUUGGUGUUUUGGCCAUCAAGCAGAUCAGCAAACCCAUUGCCAAUGUGCUGA ----------.....(((((((((.(..--..((..((((..(((((((.....))).)))))))))).).)))))))))...((((.((..((((.....))))..)).)))).. ( -31.60, z-score = -0.63, R) >droWil1.scaffold_181108 3977997 104 + 4707319 ------------AUUAGUUUUAUAAUAAUUUAAAAUGGUUAUCGGCGUAUUUCCGGCGGCCAAGUUGGGCGUUUUGGCCAUCAAACAAAUCAGCAAACCCAUUGCCAAUGUGAUCA ------------....((((((........))))))((((((.(((((.......))(((((((........)))))))........................)))...)))))). ( -20.40, z-score = 0.85, R) >droVir3.scaffold_13047 13988064 95 - 19223366 --------------------AACACCGUA-AGAAAUGGUUAUUGGAGUAUUUCCAGCCGCCAAACUUGGCGUGUUGGCAGUCAAACAAAUUAGCAAACCCAUUGCAAAUGUGUUAA --------------------...(((((.-....))))).......(.(((.((((((((((....))))).))))).)))).((((.(((.((((.....)))).))).)))).. ( -24.40, z-score = -1.14, R) >droMoj3.scaffold_6540 8050502 95 + 34148556 --------------------AUUAAUAUA-AGAGAUGGUUAUUGGAGUAUUUCCAGCCGCCAAACUUGGCGUGCUGGCAAUUAAACAAAUUAGCAAGCCCAUUGCCAAUGUUAUAA --------------------.....((((-(.(..((((...(((.((.....(((((((((....))))).))))((((((.....)))).))..)))))..)))).).))))). ( -27.80, z-score = -2.55, R) >droGri2.scaffold_14830 299483 95 + 6267026 --------------------AACAGCUUA-AGAGAUGGUUAUUGGAGUGUUUCCCGCUGCCAAGCUUGGUGCGCUGGCCAUCAAACAGAUUAGCAAGCCCAUUGCUAAUGUGCUGA --------------------..((((...-...((((((((((((((((.....)))).))))((.....))..))))))))....(.((((((((.....)))))))).))))). ( -33.00, z-score = -1.76, R) >anoGam1.chr2R 4193322 116 - 62725911 GUUAGCAAAUCCUUCGAGUUCGCCAAGCGCCAUCAUGGUUGUGGGAGCGUUUCCGGCCGCCAAGCUUGGCGUGCUGGCCAUGAAACAGAUCUCCAAACCGAUCGCCAAUCUGCUCA ...............((((..((((.((((((.(.((((.((.((((...)))).)).)))).)..))))))..)))).........((((........))))........)))). ( -36.50, z-score = -0.36, R) >triCas2.ChLG7 12183192 103 - 17478683 -------------UUAAGUAAUUAAUUUGUAAGGAUGGUAAUUCAUGCUUUCCCUGCCGCCAAACUUGGAGCCCUCUUAAUAAAACAGAUUAGUAAACCGAUCGCUAAUGCUGUCA -------------.......((((((((((..(((.((((.....)))).)))..((..(((....))).))............)))))))))).....(((.((....)).))). ( -20.40, z-score = -1.01, R) >consensus _____________UUAGUAAAAAAGUAA__AACAAUGGUUAUUGGCGUCUUUCCGGCGGCCAAGCUUGGCGUCUUGGCCAUCAAGCAGAUCAGCAAGCCGAUUGCCAAUGUGAUCA ...................................((((..((((.......))))..)))).....(((...((((............))))...)))................. ( -9.77 = -9.96 + 0.19)

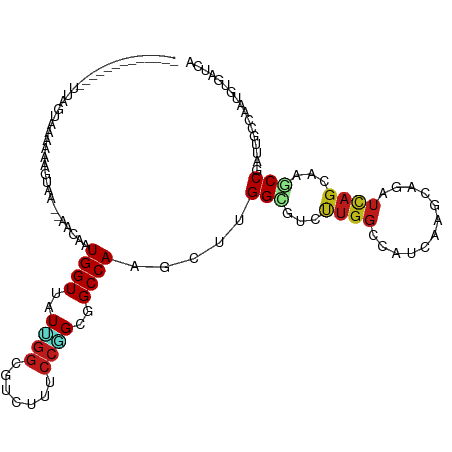
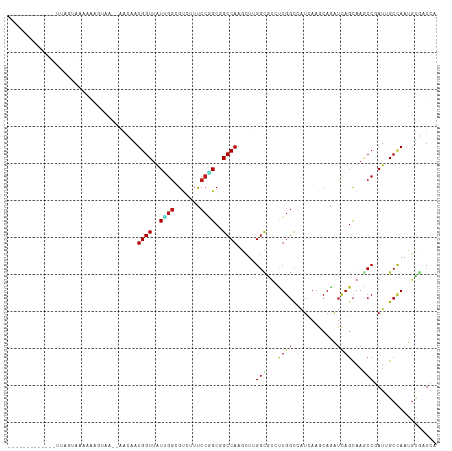
| Location | 19,753,209 – 19,753,319 |
|---|---|
| Length | 110 |
| Sequences | 14 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 66.85 |
| Shannon entropy | 0.71906 |
| G+C content | 0.45834 |
| Mean single sequence MFE | -28.13 |
| Consensus MFE | -12.40 |
| Energy contribution | -11.59 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.65 |
| Mean z-score | -1.03 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.856773 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
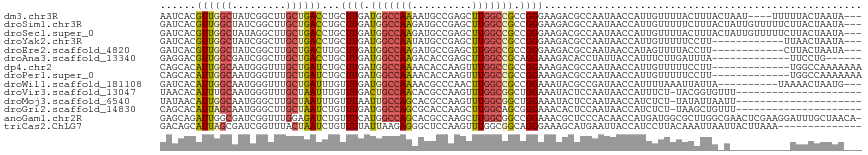
>dm3.chr3R 19753209 110 + 27905053 AAUCACGUUGGCUAUCGGCUUGCUGACCUGCUUGAUGGCCAAAAUGCCGAGCUUGGCCGCCGGGAAGACGCCAAUAACCAUUGUUUUACUUUACUAAU----UUUUUACUAAUA--- ......((((((..((((....)))).((.((((.(((((((..........))))))).)))).))..)))))).......................----............--- ( -25.20, z-score = -0.35, R) >droSim1.chr3R 19577288 114 + 27517382 GAUCACGUUGGCUAUCGGCUUGCUGACCUGCUUGAUGGCCAAGAUGCCGAGCUUGGCCGCCGGGAAGACGCCAAUAACCAUUGUUUUUCUUUACUAUUGUUUUUCUUACUAAUA--- ((..(((.(((.(((.(((...((..((.((.....(((((((........))))))))).))..))..))).))).))).)))...)).........................--- ( -29.70, z-score = -1.11, R) >droSec1.super_0 20094951 114 + 21120651 GAUCACGUUGGCUAUAGGCUUGCUGACCUGCUUGAUGGCCAAGAUGCCGAGCUUGGCCGCCGGGAAGACGCCAAUAACCAUUGUUUUACUUUACUAUUGUUUUUCUUACUAAUA--- ....(((.(((.(((.(((...((..((.((.....(((((((........))))))))).))..))..))).))).))).)))..............................--- ( -29.00, z-score = -0.99, R) >droYak2.chr3R 20646943 102 + 28832112 GAUCACGUUGGCUAUCGGCUUGCUGACCUGCUUGAUGGCCAAUAUGCCGAGCUUGGCCGCCGGGAAGACGCCAAUAACCAUUGUUUUUCCUU------------UUAACUAAUA--- ......(((((((((((((..........)).)))))))))))..((((....))))....(((((((((...........)))))))))..------------..........--- ( -28.00, z-score = -1.05, R) >droEre2.scaffold_4820 2165323 102 - 10470090 GAUCACGUUGGCUAUCGGCUUGCUGACUUGCUUGAUGGCCAAGAUGCCGAGCUUGGCCGCCGGGAAGACGCCAAUAACCAUAGUUUUACCUU------------CUUACUAAUA--- ......((((((..((((....))))(((.((((.((((((((........)))))))).)))))))..)))))).....((((........------------...))))...--- ( -30.20, z-score = -1.52, R) >droAna3.scaffold_13340 15890400 98 - 23697760 GAGGACGUUGGCGAUCGGCUUGCUGACCUGCUUGAUGGCCAAGACACCGAGCUUGGCCGCAGGAAAGACACCUAUUACCAUUUCUUGAUUUA-------------UUCCUG------ (((((...(((..((.((..((((..(((((.....(((((((........))))))))))))..)).)))).))..))).)))))......-------------......------ ( -29.70, z-score = -2.11, R) >dp4.chr2 16898491 104 - 30794189 CAGCACAUUGGCAAUGGGUUUGCUGAUCUGCUUGAUGGCCAAAACACCAAGUUUGGCCGCCGGAAAGACGCCAAUAACCAUUGUUUUUCCUU-------------UGGCCAAAAAAA .((((.((..((((.....))))..)).))))...(((........)))..((((((((..(((((((((...........)))))))))..-------------)))))))).... ( -34.40, z-score = -2.02, R) >droPer1.super_0 6549431 104 + 11822988 CAGCACAUUGGCAAUGGGUUUGCUGAUCUGCUUGAUGGCCAAAACACCAAGUUUGGCCGCCGGAAAGACGCCAAUAACCAUUGUUUUUCCUU-------------UGGCCAAAAAAA .((((.((..((((.....))))..)).))))...(((........)))..((((((((..(((((((((...........)))))))))..-------------)))))))).... ( -34.40, z-score = -2.02, R) >droWil1.scaffold_181108 3977998 104 - 4707319 GAUCACAUUGGCAAUGGGUUUGCUGAUUUGUUUGAUGGCCAAAACGCCCAACUUGGCCGCCGGAAAUACGCCGAUAACCAUUUUAAAUUAUUA----------UAAAACUAAUG--- ......(((((((((.((....)).)))..((((.(((((((..........))))))).)))).....))))))..................----------...........--- ( -20.40, z-score = 0.16, R) >droVir3.scaffold_13047 13988065 95 + 19223366 UAACACAUUUGCAAUGGGUUUGCUAAUUUGUUUGACUGCCAACACGCCAAGUUUGGCGGCUGGAAAUACUCCAAUAACCAUUUCU-UACGGUGUUU--------------------- .(((((....(.(((((..................(((((((.((.....))))))))).((((.....))))....))))).).-....))))).--------------------- ( -22.70, z-score = -0.74, R) >droMoj3.scaffold_6540 8050503 95 - 34148556 UAUAACAUUGGCAAUGGGCUUGCUAAUUUGUUUAAUUGCCAGCACGCCAAGUUUGGCGGCUGGAAAUACUCCAAUAACCAUCUCU-UAUAUUAAUU--------------------- .((((.((((((((.....))))))))))))...(((.(((((.(((((....)))))))))).)))..................-..........--------------------- ( -24.10, z-score = -1.60, R) >droGri2.scaffold_14830 299484 95 - 6267026 CAGCACAUUAGCAAUGGGCUUGCUAAUCUGUUUGAUGGCCAGCGCACCAAGCUUGGCAGCGGGAAACACUCCAAUAACCAUCUCU-UAAGCUGUUU--------------------- ((((..((((((((.....))))))))......(((((((((.((.....)))))))....(((.....)))......))))...-...))))...--------------------- ( -26.70, z-score = -0.41, R) >anoGam1.chr2R 4193323 116 + 62725911 GAGCAGAUUGGCGAUCGGUUUGGAGAUCUGUUUCAUGGCCAGCACGCCAAGCUUGGCGGCCGGAAACGCUCCCACAACCAUGAUGGCGCUUGGCGAACUCGAAGGAUUUGCUAACA- .((((((((..(((..((((..((((....))))..))))....((((((((.(((.(((.(....)))).)))...((.....)).))))))))...)))...))))))))....- ( -41.50, z-score = -1.15, R) >triCas2.ChLG7 12183193 103 + 17478683 GACAGCAUUAGCGAUCGGUUUACUAAUCUGUUUUAUUAAGAGGGCUCCAAGUUUGGCGGCAGGGAAAGCAUGAAUUACCAUCCUUACAAAUUAAUUACUUAAA-------------- (((((.(((((..(.....)..))))))))))...(((((...((((((....))).)))(((((...............)))))............))))).-------------- ( -17.76, z-score = 0.48, R) >consensus GAUCACAUUGGCAAUCGGCUUGCUGAUCUGCUUGAUGGCCAAGACGCCAAGCUUGGCCGCCGGAAAGACGCCAAUAACCAUUGUUUUACUUUA_U__________UUACUAAUA___ ......((((((.........))))))...((((.(((((((..........))))))).))))..................................................... (-12.40 = -11.59 + -0.80)


| Location | 19,753,209 – 19,753,319 |
|---|---|
| Length | 110 |
| Sequences | 14 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 66.85 |
| Shannon entropy | 0.71906 |
| G+C content | 0.45834 |
| Mean single sequence MFE | -29.45 |
| Consensus MFE | -9.77 |
| Energy contribution | -9.96 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.648668 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 19753209 110 - 27905053 ---UAUUAGUAAAAA----AUUAGUAAAGUAAAACAAUGGUUAUUGGCGUCUUCCCGGCGGCCAAGCUCGGCAUUUUGGCCAUCAAGCAGGUCAGCAAGCCGAUAGCCAACGUGAUU ---(((((((.....----)))))))........((.(((((((((((..((..((.(((((((((........))))))).....)).))..))...)))))))))))...))... ( -33.30, z-score = -2.52, R) >droSim1.chr3R 19577288 114 - 27517382 ---UAUUAGUAAGAAAAACAAUAGUAAAGAAAAACAAUGGUUAUUGGCGUCUUCCCGGCGGCCAAGCUCGGCAUCUUGGCCAUCAAGCAGGUCAGCAAGCCGAUAGCCAACGUGAUC ---...............................((.(((((((((((..((..((.(((((((((........))))))).....)).))..))...)))))))))))...))... ( -33.30, z-score = -2.49, R) >droSec1.super_0 20094951 114 - 21120651 ---UAUUAGUAAGAAAAACAAUAGUAAAGUAAAACAAUGGUUAUUGGCGUCUUCCCGGCGGCCAAGCUCGGCAUCUUGGCCAUCAAGCAGGUCAGCAAGCCUAUAGCCAACGUGAUC ---...............................((.(((((((.(((..((..((.(((((((((........))))))).....)).))..))...))).)))))))...))... ( -29.50, z-score = -1.40, R) >droYak2.chr3R 20646943 102 - 28832112 ---UAUUAGUUAA------------AAGGAAAAACAAUGGUUAUUGGCGUCUUCCCGGCGGCCAAGCUCGGCAUAUUGGCCAUCAAGCAGGUCAGCAAGCCGAUAGCCAACGUGAUC ---..........------------.........((.((((((((((((((.....)))((((..(((.(((......)))....))).)))).....)))))))))))...))... ( -31.20, z-score = -1.74, R) >droEre2.scaffold_4820 2165323 102 + 10470090 ---UAUUAGUAAG------------AAGGUAAAACUAUGGUUAUUGGCGUCUUCCCGGCGGCCAAGCUCGGCAUCUUGGCCAUCAAGCAAGUCAGCAAGCCGAUAGCCAACGUGAUC ---..........------------..(((...((..((((((((((((((.....)))(((((((........))))))).....((......))..)))))))))))..)).))) ( -33.10, z-score = -2.20, R) >droAna3.scaffold_13340 15890400 98 + 23697760 ------CAGGAA-------------UAAAUCAAGAAAUGGUAAUAGGUGUCUUUCCUGCGGCCAAGCUCGGUGUCUUGGCCAUCAAGCAGGUCAGCAAGCCGAUCGCCAACGUCCUC ------.((((.-------------............(((..((.(((..((..(((((((((((((.....).))))))).....)))))..))...))).))..)))...)))). ( -31.19, z-score = -2.20, R) >dp4.chr2 16898491 104 + 30794189 UUUUUUUGGCCA-------------AAGGAAAAACAAUGGUUAUUGGCGUCUUUCCGGCGGCCAAACUUGGUGUUUUGGCCAUCAAGCAGAUCAGCAAACCCAUUGCCAAUGUGCUG ......((((((-------------(((.(....((..((((..(((((((.....))).)))))))))).).)))))))))...((((.((..((((.....))))..)).)))). ( -31.60, z-score = -0.87, R) >droPer1.super_0 6549431 104 - 11822988 UUUUUUUGGCCA-------------AAGGAAAAACAAUGGUUAUUGGCGUCUUUCCGGCGGCCAAACUUGGUGUUUUGGCCAUCAAGCAGAUCAGCAAACCCAUUGCCAAUGUGCUG ......((((((-------------(((.(....((..((((..(((((((.....))).)))))))))).).)))))))))...((((.((..((((.....))))..)).)))). ( -31.60, z-score = -0.87, R) >droWil1.scaffold_181108 3977998 104 + 4707319 ---CAUUAGUUUUA----------UAAUAAUUUAAAAUGGUUAUCGGCGUAUUUCCGGCGGCCAAGUUGGGCGUUUUGGCCAUCAAACAAAUCAGCAAACCCAUUGCCAAUGUGAUC ---.....((((((----------........))))))(((..((((.......))))..)))..(((((((......))).............((((.....))))))))...... ( -19.60, z-score = 0.91, R) >droVir3.scaffold_13047 13988065 95 - 19223366 ---------------------AAACACCGUA-AGAAAUGGUUAUUGGAGUAUUUCCAGCCGCCAAACUUGGCGUGUUGGCAGUCAAACAAAUUAGCAAACCCAUUGCAAAUGUGUUA ---------------------....(((((.-....))))).......(.(((.((((((((((....))))).))))).)))).((((.(((.((((.....)))).))).)))). ( -24.40, z-score = -1.14, R) >droMoj3.scaffold_6540 8050503 95 + 34148556 ---------------------AAUUAAUAUA-AGAGAUGGUUAUUGGAGUAUUUCCAGCCGCCAAACUUGGCGUGCUGGCAAUUAAACAAAUUAGCAAGCCCAUUGCCAAUGUUAUA ---------------------......((((-(.(..((((...(((.((.....(((((((((....))))).))))((((((.....)))).))..)))))..)))).).))))) ( -27.00, z-score = -2.28, R) >droGri2.scaffold_14830 299484 95 + 6267026 ---------------------AAACAGCUUA-AGAGAUGGUUAUUGGAGUGUUUCCCGCUGCCAAGCUUGGUGCGCUGGCCAUCAAACAGAUUAGCAAGCCCAUUGCUAAUGUGCUG ---------------------...((((...-...((((((((((((((((.....)))).))))((.....))..))))))))....(.((((((((.....)))))))).))))) ( -31.90, z-score = -1.62, R) >anoGam1.chr2R 4193323 116 - 62725911 -UGUUAGCAAAUCCUUCGAGUUCGCCAAGCGCCAUCAUGGUUGUGGGAGCGUUUCCGGCCGCCAAGCUUGGCGUGCUGGCCAUGAAACAGAUCUCCAAACCGAUCGCCAAUCUGCUC -....((((.........(((.((((((((.......((((.((.((((...)))).)).)))).)))))))).)))(((..((...))((((........)))))))....)))). ( -35.90, z-score = -0.23, R) >triCas2.ChLG7 12183193 103 - 17478683 --------------UUUAAGUAAUUAAUUUGUAAGGAUGGUAAUUCAUGCUUUCCCUGCCGCCAAACUUGGAGCCCUCUUAAUAAAACAGAUUAGUAAACCGAUCGCUAAUGCUGUC --------------........((((((((((..(((.((((.....)))).)))..((..(((....))).))............)))))))))).....(((.((....)).))) ( -18.70, z-score = -0.53, R) >consensus ___UAUUAGUAA__________A_UAAAGUAAAACAAUGGUUAUUGGCGUCUUUCCGGCGGCCAAGCUUGGCGUCUUGGCCAUCAAGCAGAUCAGCAAGCCGAUUGCCAAUGUGAUC .....................................((((..((((.......))))..)))).....(((...((((............))))...)))................ ( -9.77 = -9.96 + 0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:39:14 2011