
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,748,001 – 19,748,103 |
| Length | 102 |
| Max. P | 0.794550 |

| Location | 19,748,001 – 19,748,102 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 97.43 |
| Shannon entropy | 0.04535 |
| G+C content | 0.59010 |
| Mean single sequence MFE | -34.95 |
| Consensus MFE | -33.98 |
| Energy contribution | -34.02 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.97 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.794550 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
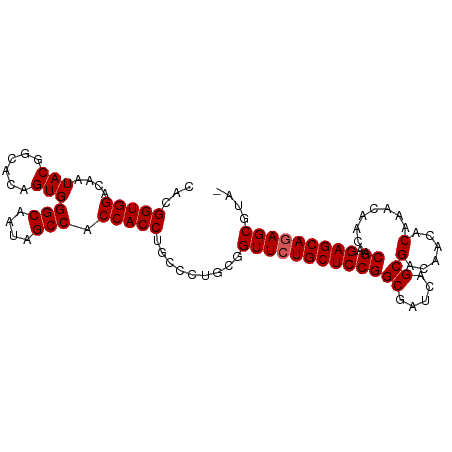
>dm3.chr3R 19748001 101 + 27905053 UGCACGGUGGACAAUACGGCACAGUGGGCAAUAGCCACCACCUGCCCUGCGGUUCUGCUCCGGCGAUCAGCGACAACACAAACAACACGAGAGCAGAGCGU .(((.(((((....(((......)))(((....))).))))))))......(((((((((((((.....))(......)........)).))))))))).. ( -35.20, z-score = -1.46, R) >droEre2.scaffold_4820 2160076 101 - 10470090 UGCGCGGUGGACAAUACGACACAGUGGGCAAUAGCCACCACCUGCCCUGUGGUUCUGCUCCGGCGAUCAGCGACAACGCAAACAACACGAGAGCAGAGCCU .(((.(((((....(((......)))(((....))).)))))))).....((((((((((((.......(((....)))........)).)))))))))). ( -39.66, z-score = -2.73, R) >droYak2.chr3R 20641649 101 + 28832112 UGCACGGUGGACAAUACGGCACAGUGGGCAAUAGCCACCACCUGCCCUGUGGUUCUGCUCCGGCGAUCAGCGACAACACAAACAACACGAGAGCAGAGCGU .(((.(((((....(((......)))(((....))).))))))))......(((((((((((((.....))(......)........)).))))))))).. ( -35.20, z-score = -1.34, R) >droSec1.super_0 20089756 101 + 21120651 UGCACGGUGGACAAUACGGCACAGUGGGCAAUAGCCACCACCUGCCCUGCGGUUCUGCUCCGGCGAUCAGCGACAACACAAACAACACGAGAGCAGAGCGU .(((.(((((....(((......)))(((....))).))))))))......(((((((((((((.....))(......)........)).))))))))).. ( -35.20, z-score = -1.46, R) >droSim1.chr3R 19570835 101 + 27517382 UGCACGGUGGACAAUACGGCACAGUGGGCAAUAGCCACCACCUGCCCUGCGGUUCUGCUCCGGCGAUCAGCGACAACACAAACAACACGAGAGCAAAGCGU .(((.(((((....(((......)))(((....))).))))))))...(((.((.(((((((((.....))(......)........)).))))))).))) ( -29.50, z-score = 0.11, R) >consensus UGCACGGUGGACAAUACGGCACAGUGGGCAAUAGCCACCACCUGCCCUGCGGUUCUGCUCCGGCGAUCAGCGACAACACAAACAACACGAGAGCAGAGCGU .(((.(((((....(((......)))(((....))).))))))))......(((((((((((((.....))(......)........)).))))))))).. (-33.98 = -34.02 + 0.04)


| Location | 19,748,003 – 19,748,103 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 96.41 |
| Shannon entropy | 0.06211 |
| G+C content | 0.59081 |
| Mean single sequence MFE | -33.07 |
| Consensus MFE | -30.86 |
| Energy contribution | -31.06 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr3R 19748003 100 + 27905053 CACGGUGGACAAUACGGCACAGUGGGCAAUAGCCACCACCUGCCCUGCGGUUCUGCUCCGGCGAUCAGCGACAACACAAACAACACGAGAGCAGAGCGUA- .(((((((....(((......)))(((....))).))))).........(((((((((((((.....))(......)........)).))))))))))).- ( -32.20, z-score = -1.02, R) >droEre2.scaffold_4820 2160078 100 - 10470090 CGCGGUGGACAAUACGACACAGUGGGCAAUAGCCACCACCUGCCCUGUGGUUCUGCUCCGGCGAUCAGCGACAACGCAAACAACACGAGAGCAGAGCCUC- .(((((((....(((......)))(((....))).))))).)).....((((((((((((.......(((....)))........)).))))))))))..- ( -38.86, z-score = -2.88, R) >droYak2.chr3R 20641651 101 + 28832112 CACGGUGGACAAUACGGCACAGUGGGCAAUAGCCACCACCUGCCCUGUGGUUCUGCUCCGGCGAUCAGCGACAACACAAACAACACGAGAGCAGAGCGUCC ......((((.(((.((((..((((((....))..)))).)))).))).(((((((((((((.....))(......)........)).))))))))))))) ( -34.80, z-score = -1.50, R) >droSec1.super_0 20089758 100 + 21120651 CACGGUGGACAAUACGGCACAGUGGGCAAUAGCCACCACCUGCCCUGCGGUUCUGCUCCGGCGAUCAGCGACAACACAAACAACACGAGAGCAGAGCGUA- .(((((((....(((......)))(((....))).))))).........(((((((((((((.....))(......)........)).))))))))))).- ( -32.20, z-score = -1.02, R) >droSim1.chr3R 19570837 100 + 27517382 CACGGUGGACAAUACGGCACAGUGGGCAAUAGCCACCACCUGCCCUGCGGUUCUGCUCCGGCGAUCAGCGACAACACAAACAACACGAGAGCAAAGCGUA- ...(((((....(((......)))(((....))).))))).....((((.((.(((((((((.....))(......)........)).))))))).))))- ( -27.30, z-score = 0.35, R) >consensus CACGGUGGACAAUACGGCACAGUGGGCAAUAGCCACCACCUGCCCUGCGGUUCUGCUCCGGCGAUCAGCGACAACACAAACAACACGAGAGCAGAGCGUA_ ...(((((....(((......)))(((....))).))))).........(((((((((((((.....))(......)........)).))))))))).... (-30.86 = -31.06 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:39:09 2011