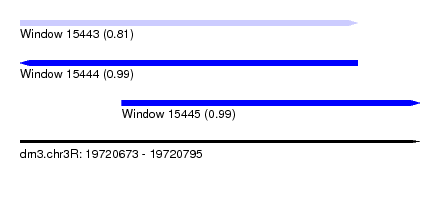
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,720,673 – 19,720,795 |
| Length | 122 |
| Max. P | 0.993654 |

| Location | 19,720,673 – 19,720,776 |
|---|---|
| Length | 103 |
| Sequences | 13 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 66.60 |
| Shannon entropy | 0.68107 |
| G+C content | 0.55233 |
| Mean single sequence MFE | -32.97 |
| Consensus MFE | -12.77 |
| Energy contribution | -12.69 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.810790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
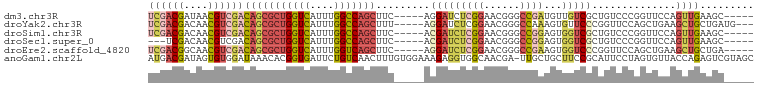
>dm3.chr3R 19720673 103 + 27905053 AUAACGAGGGUGUAGCUGGCUGUGCU---------CACUGUCGACGAUAACGUCGACAG---CGCUGGUCAUUUGGCCAGCUUCAGGAUCUCGGAACGGGCCGAUGUUGUCGCUG .......(((..(((....)))..))---------).(((((((((....)))))))))---.(((((((....)))))))..((((((.((((......))))....))).))) ( -43.00, z-score = -2.40, R) >droYak2.chr3R 20612888 88 + 28832112 AUAACGAGGGUGUAGCUGG------------------AUGUCGACGACAACGUCGACAG---CGCUGGUCAUUUGGCCAGCUUUAGGAUCUCGGAACGGGCCAAAGUGU------ ...((...(((.(..((((------------------(((((((((....)))))))..---.(((((((....)))))))......))).)))...).)))...))..------ ( -33.50, z-score = -2.44, R) >droSim1.chr3R 19541604 103 + 27517382 AUAACGAGGGUGUAGCUGGCUGUGCU---------CACUGUCGACGACAACGUCGACAG---CGCUGGUCAUUUGGCCAGCUUCACGAUCUCGGAACGGGCCGGAGUGGUCGCUG ....((((((((.(((((((((((..---------(((((((((((....)))))))))---...))..)))..)))))))).)))..))))).....((((.....)))).... ( -45.40, z-score = -2.45, R) >droSec1.super_0 20062606 100 + 21120651 AUAACGAGGGUGUAGCUGGCUGUGCU---------CACUGUCGA---CAACGUCGACAG---CGCUGGUCAUUUGGCCAGCUUCACGAUCUCGGAACGGGCCGGAGUGGUCGCUG ....((((((((.(((((((((((..---------(((((((((---(...))))))))---...))..)))..)))))))).)))..))))).....((((.....)))).... ( -41.80, z-score = -1.80, R) >droAna3.scaffold_13340 15859925 101 - 23697760 AUGACGAGCCUGUAGUUGACUGUUUGUGUUGCUGUUACCGUCGCCGACAUCGCCGACACAUCGGCUGGUCAUUUGGCCAGCUUCAGGGUUUCCGGCGAGCG-------------- ...((((((..((.....)).)))))).((((((..(((((((.((....)).)))).....((((((((....))))))))....)))...))))))...-------------- ( -35.50, z-score = -0.70, R) >droEre2.scaffold_4820 2132246 97 - 10470090 AUAACGAGGGUGUAGCUGGCUGUGCU---------CACUGUCGACGGCAACGUCGACAG---CGCUGGUCAUUUGGUCAGCUUCAGGAUCUCGGAACGGGCCGAAGUGG------ .......(((..(((....)))..))---------).(((((((((....)))))))))---.((((..(....)..)))).........((((......)))).....------ ( -41.10, z-score = -2.59, R) >dp4.chr2 16864691 82 - 30794189 AUGACGAGCGUGUAGUUCGCUGUGC---------UUACUGUCGACGACAACGCCGACAG---CGCAGGUCAUUUGGCCAGCUUCAGGAUCUCCG--------------------- .(((.((((.....))))(((.(((---------...((((((.((....)).))))))---.)))((((....))))))).)))((....)).--------------------- ( -26.80, z-score = -0.36, R) >droPer1.super_0 6514782 82 + 11822988 AUGACGAGCGUGUAGUUCGCUGUGC---------UUACUGUCGACGACAACGCCGACAA---CGCAGGUCAUUUGGCCAGCUUCAGGAUCUCCG--------------------- .(((.((((.....))))(((.(((---------....(((((.((....)).))))).---.)))((((....))))))).)))((....)).--------------------- ( -23.70, z-score = 0.19, R) >droWil1.scaffold_181108 3943869 106 - 4707319 AUGAUGAGCGUAUAGUUUGCUGUGGUGGUGG---UUAAUGUCGACGAGAACGUCGACAA---UUCCGGUCAUUUGGUCAGCUUCAAGAUAUCUGCCUUGCCAUGUGGAAGUA--- ...............((..(.(((((((..(---.((.((((((((....)))))))).---....(..(....)..)..........)).)..))..))))))..))....--- ( -27.80, z-score = 0.10, R) >droVir3.scaffold_13047 13957966 80 + 19223366 AUAACGAGCGUAUAGUUUGCUGUGGU---------UACUGUCGACGGCAGCGCCGACAG---CGCUGGUCAUUUGGCCAGCUUCAGGAUCUC----------------------- .....((((.....))))((((((((---------..((((.....)))).))).))))---)(((((((....)))))))...........----------------------- ( -29.20, z-score = -1.56, R) >droMoj3.scaffold_6540 8020342 80 - 34148556 AUGACGAGCGUAUAGUUUGCUGUGGUG---------ACUGUCGACGGCAACGCCGACAG---CGCUGGUCAUUUGGCCAGCUUCAGGAUCUC----------------------- ....(((((.....)))))(((....(---------.((((((.((....)).))))))---)(((((((....)))))))..)))......----------------------- ( -31.50, z-score = -1.96, R) >droGri2.scaffold_15074 7652464 82 - 7742996 AUAACGAGCGUAUAGUUUGCUGUGGU---------UAUUGUCGACGGCAACGCCGCCAG---CGCUGGUCAUUUGGCCAGCUUCAAGAUCUCCG--------------------- .....((((.....))))((((((((---------..((((.....)))).)))).)))---)(((((((....))))))).............--------------------- ( -26.90, z-score = -1.16, R) >anoGam1.chr2L 1013958 95 + 48795086 AUAUCGAUGAUGUAGAUGCCGAUGUG-------------GAUGACGAUAGUGUGGAUAA---ACACGGUGAUUCUGUCAACUUUGUGGAAAGAGGUGGCAACGAUUGCUGC---- .(((((.(.((.((.((....)).))-------------.)).))))))((((......---))))((..(((.(((((.((((......)))).)))))..)))..))..---- ( -22.40, z-score = -0.56, R) >consensus AUAACGAGCGUGUAGUUGGCUGUGCU_________UACUGUCGACGACAACGUCGACAG___CGCUGGUCAUUUGGCCAGCUUCAGGAUCUCGG___GG_C______________ .....(((.....((((.....................(((((.((....)).)))))........((((....)))))))).......)))....................... (-12.77 = -12.69 + -0.07)

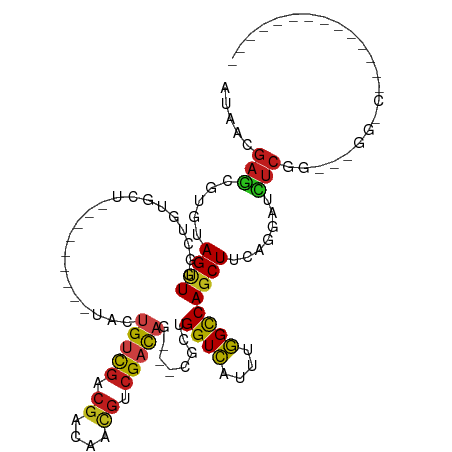
| Location | 19,720,673 – 19,720,776 |
|---|---|
| Length | 103 |
| Sequences | 13 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 66.60 |
| Shannon entropy | 0.68107 |
| G+C content | 0.55233 |
| Mean single sequence MFE | -28.35 |
| Consensus MFE | -14.13 |
| Energy contribution | -15.09 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.63 |
| SVM RNA-class probability | 0.993654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
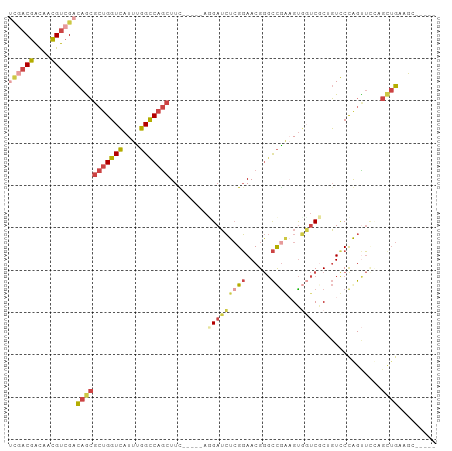
>dm3.chr3R 19720673 103 - 27905053 CAGCGACAACAUCGGCCCGUUCCGAGAUCCUGAAGCUGGCCAAAUGACCAGCG---CUGUCGACGUUAUCGUCGACAGUG---------AGCACAGCCAGCUACACCCUCGUUAU .(((((.....((((......)))).....((.(((((((....((.....((---(((((((((....)))))))))))---------..))..))))))).))...))))).. ( -40.10, z-score = -4.21, R) >droYak2.chr3R 20612888 88 - 28832112 ------ACACUUUGGCCCGUUCCGAGAUCCUAAAGCUGGCCAAAUGACCAGCG---CUGUCGACGUUGUCGUCGACAU------------------CCAGCUACACCCUCGUUAU ------.((.(((((((.(((...((...))..))).)))))))))...(((.---.((((((((....)))))))).------------------...)))............. ( -26.80, z-score = -2.61, R) >droSim1.chr3R 19541604 103 - 27517382 CAGCGACCACUCCGGCCCGUUCCGAGAUCGUGAAGCUGGCCAAAUGACCAGCG---CUGUCGACGUUGUCGUCGACAGUG---------AGCACAGCCAGCUACACCCUCGUUAU .(((((...(((.((......)))))...(((.(((((((....((.....((---(((((((((....)))))))))))---------..))..))))))).)))..))))).. ( -42.60, z-score = -3.95, R) >droSec1.super_0 20062606 100 - 21120651 CAGCGACCACUCCGGCCCGUUCCGAGAUCGUGAAGCUGGCCAAAUGACCAGCG---CUGUCGACGUUG---UCGACAGUG---------AGCACAGCCAGCUACACCCUCGUUAU .(((((...(((.((......)))))...(((.(((((((....((.....((---((((((((...)---)))))))))---------..))..))))))).)))..))))).. ( -38.60, z-score = -3.30, R) >droAna3.scaffold_13340 15859925 101 + 23697760 --------------CGCUCGCCGGAAACCCUGAAGCUGGCCAAAUGACCAGCCGAUGUGUCGGCGAUGUCGGCGACGGUAACAGCAACACAAACAGUCAACUACAGGCUCGUCAU --------------.....(((((...........)))))...(((((.((((..(((((..((..(..((....))..)...)).)))))...((....))...)))).))))) ( -28.70, z-score = 0.26, R) >droEre2.scaffold_4820 2132246 97 + 10470090 ------CCACUUCGGCCCGUUCCGAGAUCCUGAAGCUGACCAAAUGACCAGCG---CUGUCGACGUUGCCGUCGACAGUG---------AGCACAGCCAGCUACACCCUCGUUAU ------.....((((......))))((...((.(((((.............((---(((((((((....)))))))))))---------.((...))))))).))...))..... ( -29.60, z-score = -2.07, R) >dp4.chr2 16864691 82 + 30794189 ---------------------CGGAGAUCCUGAAGCUGGCCAAAUGACCUGCG---CUGUCGGCGUUGUCGUCGACAGUAA---------GCACAGCGAACUACACGCUCGUCAU ---------------------.((....)).............(((((.((((---(((((((((....))))))))))..---------))).((((.......)))).))))) ( -26.70, z-score = -0.56, R) >droPer1.super_0 6514782 82 - 11822988 ---------------------CGGAGAUCCUGAAGCUGGCCAAAUGACCUGCG---UUGUCGGCGUUGUCGUCGACAGUAA---------GCACAGCGAACUACACGCUCGUCAU ---------------------.((....)).............(((((..(((---(.(((((((....)))))))(((..---------((...))..)))..))))..))))) ( -24.10, z-score = 0.09, R) >droWil1.scaffold_181108 3943869 106 + 4707319 ---UACUUCCACAUGGCAAGGCAGAUAUCUUGAAGCUGACCAAAUGACCGGAA---UUGUCGACGUUCUCGUCGACAUUAA---CCACCACCACAGCAAACUAUACGCUCAUCAU ---.........(((((((((......)))))..((((.((........))..---.((((((((....))))))))....---.........))))...))))........... ( -23.20, z-score = -1.78, R) >droVir3.scaffold_13047 13957966 80 - 19223366 -----------------------GAGAUCCUGAAGCUGGCCAAAUGACCAGCG---CUGUCGGCGCUGCCGUCGACAGUA---------ACCACAGCAAACUAUACGCUCGUUAU -----------------------(((........(((((.(....).)))))(---(((((((((....)))))))))).---------..................)))..... ( -27.20, z-score = -2.41, R) >droMoj3.scaffold_6540 8020342 80 + 34148556 -----------------------GAGAUCCUGAAGCUGGCCAAAUGACCAGCG---CUGUCGGCGUUGCCGUCGACAGU---------CACCACAGCAAACUAUACGCUCGUCAU -----------------------(((........(((((.(....).)))))(---(((((((((....))))))))))---------...................)))..... ( -26.90, z-score = -2.05, R) >droGri2.scaffold_15074 7652464 82 + 7742996 ---------------------CGGAGAUCUUGAAGCUGGCCAAAUGACCAGCG---CUGGCGGCGUUGCCGUCGACAAUA---------ACCACAGCAAACUAUACGCUCGUUAU ---------------------..(((........(((((.(....).)))))(---((((((((...))))).(......---------..).))))..........)))..... ( -22.10, z-score = -0.18, R) >anoGam1.chr2L 1013958 95 - 48795086 ----GCAGCAAUCGUUGCCACCUCUUUCCACAAAGUUGACAGAAUCACCGUGU---UUAUCCACACUAUCGUCAUC-------------CACAUCGGCAUCUACAUCAUCGAUAU ----......((((.((((....((((....)))).((((.(......)((((---......))))....))))..-------------......))))..........)))).. ( -11.90, z-score = -0.28, R) >consensus ______________G_CC___CCGAGAUCCUGAAGCUGGCCAAAUGACCAGCG___CUGUCGACGUUGUCGUCGACAGUA_________AGCACAGCCAACUACACGCUCGUUAU .......................(((........((((..........)))).....((((((((....))))))))..............................)))..... (-14.13 = -15.09 + 0.97)

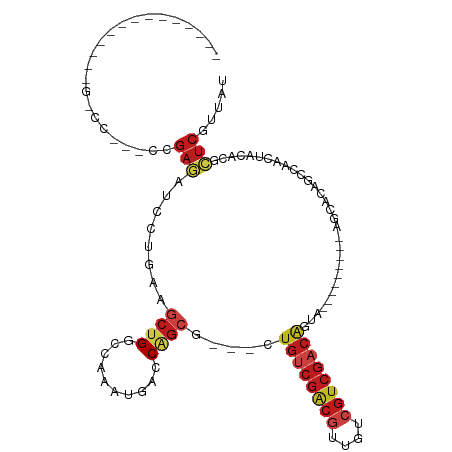
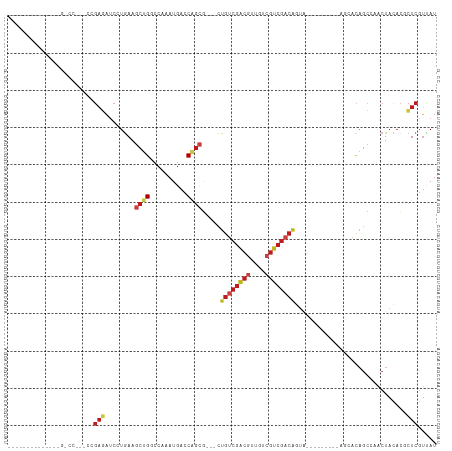
| Location | 19,720,704 – 19,720,795 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 70.98 |
| Shannon entropy | 0.51994 |
| G+C content | 0.57595 |
| Mean single sequence MFE | -38.12 |
| Consensus MFE | -18.67 |
| Energy contribution | -20.15 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.985982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
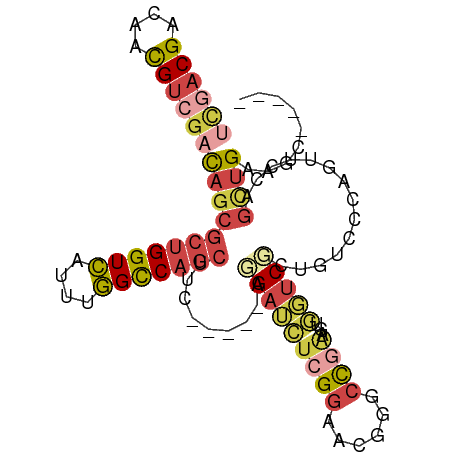
>dm3.chr3R 19720704 91 + 27905053 UCGACGAUAACGUCGACAGCGCUGGUCAUUUGGCCAGCUUC-----AGGAUCUCGGAACGGGCCGAUGUUGUCGCUGUCCCGGUUCCAGUUGAAGC----- ((((((....))))))(((((((((((....)))))))...-----.(((.(.(((.((((..((((...)))))))).)))).))).))))....----- ( -37.60, z-score = -2.23, R) >droYak2.chr3R 20612910 93 + 28832112 UCGACGACAACGUCGACAGCGCUGGUCAUUUGGCCAGCUUU-----AGGAUCUCGGAACGGGCCAAAGUGUUCCCGGUUCCAGCUGAAGCUGCUGAUG--- ((((((....))))))(((((((((((....)))))))...-----.(((.((.((((((........)))))).)).)))(((....)))))))...--- ( -40.40, z-score = -3.32, R) >droSim1.chr3R 19541635 91 + 27517382 UCGACGACAACGUCGACAGCGCUGGUCAUUUGGCCAGCUUC-----ACGAUCUCGGAACGGGCCGGAGUGGUCGCUGUCCCGGUUCCAGUUGAAGC----- ((((((....))))))(((((((((((....)))))))...-----........((((((((.(((.(....).))).))).))))).))))....----- ( -39.10, z-score = -1.94, R) >droSec1.super_0 20062637 88 + 21120651 ---UCGACAACGUCGACAGCGCUGGUCAUUUGGCCAGCUUC-----ACGAUCUCGGAACGGGCCGGAGUGGUCGCUGUCCCGGUUCCAGUUGAAGC----- ---(((((...)))))(((((((((((....)))))))...-----........((((((((.(((.(....).))).))).))))).))))....----- ( -35.50, z-score = -1.36, R) >droEre2.scaffold_4820 2132277 91 - 10470090 UCGACGGCAACGUCGACAGCGCUGGUCAUUUGGUCAGCUUC-----AGGAUCUCGGAACGGGCCGAAGUGGUCCCGGUUCCAGCUGAAGCUGCUGA----- ((((((....))))))((((...(..(....)..)((((((-----((...((.((((((((((.....)).))).))))))))))))))))))).----- ( -44.60, z-score = -3.43, R) >anoGam1.chr2L 1013985 100 + 48795086 AUGACGAUAGUGUGGAUAAACACGGUGAUUCUGUCAACUUUGUGGAAAGAGGUGGCAACGA-UUGCUGCUUCCGCAUUCCUAGUGUUACCAGAGUCGUAGC ...(((((.((((......))))((((((.(((.......((((((....((..(((....-.)))..))))))))....))).))))))...)))))... ( -31.50, z-score = -1.84, R) >consensus UCGACGACAACGUCGACAGCGCUGGUCAUUUGGCCAGCUUC_____AGGAUCUCGGAACGGGCCGAAGUGGUCGCUGUCCCAGUUCCAGCUGAAGC_____ ((((((....))))))(((((((((((....))))))).........(((((((((......))))...)))))..............))))......... (-18.67 = -20.15 + 1.48)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:39:04 2011