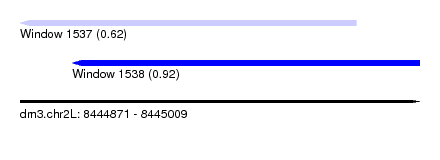
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,444,871 – 8,445,009 |
| Length | 138 |
| Max. P | 0.920008 |

| Location | 8,444,871 – 8,444,987 |
|---|---|
| Length | 116 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.85 |
| Shannon entropy | 0.63701 |
| G+C content | 0.48715 |
| Mean single sequence MFE | -28.91 |
| Consensus MFE | -11.56 |
| Energy contribution | -11.20 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.85 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.618842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
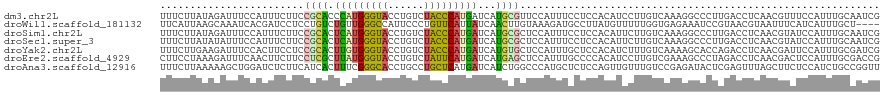
>dm3.chr2L 8444871 116 - 23011544 UCCGCACCCAUGGGUACCUGUCUACCCAUGAUCAUGCGUUCCAUUUCCUCCACAUCCUUGUCAAAGGCCCUUGACCUCAACGUUUCCAUUUGCAAUCGCAGAAGGGUGUCAUGCAU---- ..((((..((((((((......))))))))....)))).............(((((((((((((......)))))..............((((....)))))))))))).......---- ( -30.70, z-score = -1.86, R) >droWil1.scaffold_181132 337779 112 - 1035393 UCCUGUCUGUUGGGCCAUUCCCUGUUCAUUAUCAACUUGUAAAGAUGCCUUAUGUUUUUGGUGAGAAAUCCGUAACGUAAUUUCAUCAUUUGCU-UGGUAAAGAAACGUC-UAU------ ...........(((.....)))..(((.(((((((...(((((((((..(((((((..(((........))).)))))))...)))).))))))-)))))).))).....-...------ ( -22.00, z-score = -0.83, R) >droSim1.chr2L 8231208 116 - 22036055 UCCGCACUCAUGGGUACCUGUCUACCCAUGAUCAUGCGCUCCAUUUCCUCCACAUUCUUGUCAAAGGCCCUUGACCUCAACGUAUCCAUUUGCAAUCGCAGUAGAGUGUCGUGCAU---- ...(((((((((((((......)))))))))....((((((..................(((((......)))))..............((((....))))..)))))).))))..---- ( -32.30, z-score = -3.03, R) >droSec1.super_3 3941384 116 - 7220098 UCCGCACUCAUGGGUACCUGUCUACCCAUGAUCAUGCGCUCCAUUUCCUCCACAUUCUUGUCAAAGGCCCUUGACCUCAACGUAUCCAUUUGCAAUCGCAGUAGGGUGUCGUGCAU---- ...(((.(((((((((......)))))))))...)))((....................(((((......)))))....((((((((..((((....))))..))))).)))))..---- ( -32.60, z-score = -2.44, R) >droYak2.chr2L 11082330 116 - 22324452 UCCGCACUUGUGGGUACCUGUCUACCCAUGAUCAUGUGCUCCAUUUGCUCCACAUCUUUGUCAAAAGCACCAGACCUCAACGAUUCCAUUUGCGAUCGGACAAAGGCGUCGUGCGU---- ...(((((..((((((......))))))..)....)))).......((.(.((..(((((((..........(....)..(((((.(....).))))))))))))..)).).))..---- ( -31.90, z-score = -1.22, R) >droEre2.scaffold_4929 17354123 116 - 26641161 UCCUCGCUUAUGGGUACCUGUCUAUUCAUGAUCAUGAGCUCCAUUUGCCCCACAUCCUUGUCGAAAGCCCUAGACCUCAACGACUCCAUUUGCGACCGAACGAAGGCGUCGUGCGU---- ....(((....(((((..((....(((((....)))))...))..))))).........(((..........)))....(((((.((.((((........)))))).)))))))).---- ( -23.40, z-score = 0.79, R) >droAna3.scaffold_12916 10099500 120 - 16180835 UCAUCACUUUCGGGCACCUGCCUGCUCAUGAUCAUCUGGCCCAUGCUCUCCAGUUGUUUGUCCGAGAUACUCGAGUUUAGCUUCUCCAUCUGCCGGUUCAGGAGCGUAUAAUGAAUAAAU ((((.......((((....))))((((.(((....(((((..(((......(((((...(..((((...))))..).)))))....)))..))))).))).)))).....))))...... ( -29.50, z-score = -0.12, R) >consensus UCCGCACUCAUGGGUACCUGUCUACCCAUGAUCAUGCGCUCCAUUUCCUCCACAUCCUUGUCAAAGGCCCUUGACCUCAACGUCUCCAUUUGCAAUCGCAGAAGGGUGUCGUGCAU____ ..((((.(((((((((......)))))))))...))))....................................................((((..(((......)))...))))..... (-11.56 = -11.20 + -0.36)
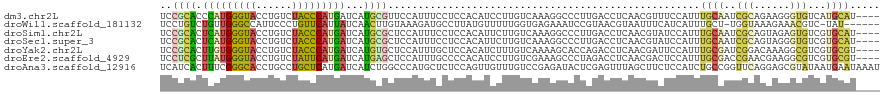
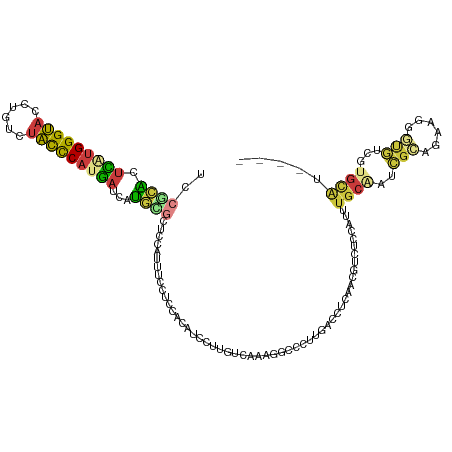
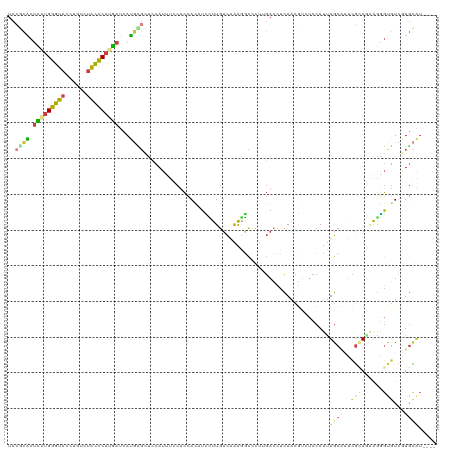
| Location | 8,444,889 – 8,445,009 |
|---|---|
| Length | 120 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.68 |
| Shannon entropy | 0.61826 |
| G+C content | 0.45062 |
| Mean single sequence MFE | -21.77 |
| Consensus MFE | -10.54 |
| Energy contribution | -9.83 |
| Covariance contribution | -0.71 |
| Combinations/Pair | 1.85 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.920008 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
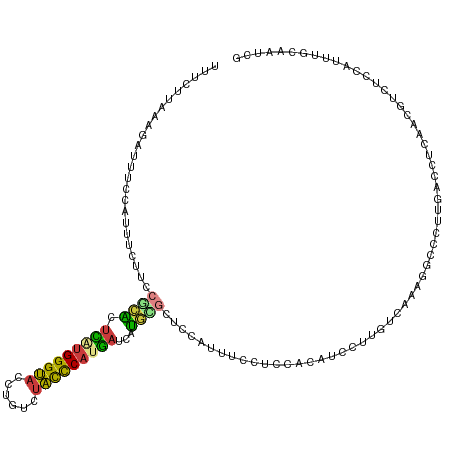
>dm3.chr2L 8444889 120 - 23011544 UUUCUUAUAGAUUUCCAUUUCUUCCGCACCCAUGGGUACCUGUCUACCCAUGAUCAUGCGUUCCAUUUCCUCCACAUCCUUGUCAAAGGCCCUUGACCUCAACGUUUCCAUUUGCAAUCG .........((((...........((((..((((((((......))))))))....)))).....................(((((......)))))..................)))). ( -20.40, z-score = -2.08, R) >droWil1.scaffold_181132 337797 116 - 1035393 UUCAUUAAGCAAAUCACGAUCCUCCUGUCUGUUGGGCCAUUCCCUGUUCAUUAUCAACUUGUAAAGAUGCCUUAUGUUUUUGGUGAGAAAUCCGUAACGUAAUUUCAUCAUUUGCU---- .......(((((((.((((..............(((.....)))..............))))...((((..(((((((..(((........))).)))))))...)))))))))))---- ( -19.39, z-score = -0.08, R) >droSim1.chr2L 8231226 120 - 22036055 UUUCUUAUAGAUUUCCAUUUCUUCCGCACUCAUGGGUACCUGUCUACCCAUGAUCAUGCGCUCCAUUUCCUCCACAUUCUUGUCAAAGGCCCUUGACCUCAACGUAUCCAUUUGCAAUCG .........((((...........((((.(((((((((......)))))))))...)))).....................(((((......)))))......(((......))))))). ( -23.00, z-score = -2.98, R) >droSec1.super_3 3941402 120 - 7220098 UUUCUUAUAUAUUUCCAUUUCUUCCGCACUCAUGGGUACCUGUCUACCCAUGAUCAUGCGCUCCAUUUCCUCCACAUUCUUGUCAAAGGCCCUUGACCUCAACGUAUCCAUUUGCAAUCG ........................((((.(((((((((......)))))))))...)))).....................(((((......)))))......(((......)))..... ( -21.80, z-score = -3.22, R) >droYak2.chr2L 11082348 120 - 22324452 UUUCUUGAAGAUUUCCACUUCCUCCGCACUUGUGGGUACCUGUCUACCCAUGAUCAUGUGCUCCAUUUGCUCCACAUCUUUGUCAAAAGCACCAGACCUCAACGAUUCCAUUUGCGAUCG ......((((.......))))...((((..((((((((......))))))(((((..(((((...((((....(((....))))))))))))..))..))).......))..)))).... ( -22.00, z-score = -0.89, R) >droEre2.scaffold_4929 17354141 120 - 26641161 CUUCCUAAAGAUUUCAACUUCUUCCUCGCUUAUGGGUACCUGUCUAUUCAUGAUCAUGAGCUCCAUUUGCCCCACAUCCUUGUCGAAAGCCCUAGACCUCAACGACUCCAUUUGCGACCG .......((((........))))..((((..((((((....(((((.((((....))))(((...((((....(((....))))))))))..))))).......)).))))..))))... ( -20.20, z-score = -1.03, R) >droAna3.scaffold_12916 10099522 120 - 16180835 UUUCUUAAAAAGCUGGAUCUCUUCAUCACUUUCGGGCACCUGCCUGCUCAUGAUCAUCUGGCCCAUGCUCUCCAGUUGUUUGUCCGAGAUACUCGAGUUUAGCUUCUCCAUCUGCCGGUU ..........(((((((((((...((((....(((((....)))))....))))((.((((..........)))).)).......)))))....(((........)))......)))))) ( -25.60, z-score = 0.43, R) >consensus UUUCUUAAAGAUUUCCAUUUCUUCCGCACUCAUGGGUACCUGUCUACCCAUGAUCAUGCGCUCCAUUUCCUCCACAUCCUUGUCAAAGGCCCUUGACCUCAACGUCUCCAUUUGCAAUCG ........................((((.(((((((((......)))))))))...))))............................................................ (-10.54 = -9.83 + -0.71)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:26:05 2011