
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,687,790 – 19,687,887 |
| Length | 97 |
| Max. P | 0.685320 |

| Location | 19,687,790 – 19,687,887 |
|---|---|
| Length | 97 |
| Sequences | 12 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 72.28 |
| Shannon entropy | 0.60719 |
| G+C content | 0.42731 |
| Mean single sequence MFE | -19.88 |
| Consensus MFE | -11.89 |
| Energy contribution | -11.48 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.33 |
| Mean z-score | -0.54 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.685320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
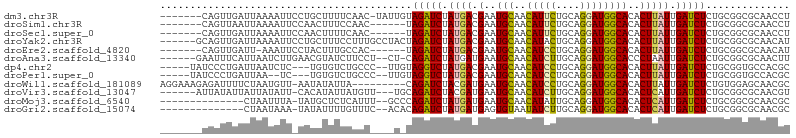
>dm3.chr3R 19687790 97 - 27905053 -------CAGUUGAUUAAAAUUCCUGCUUUUCAAC-UAUUGUAGAUCUAUGACGAAUGCAACAUUCUGCAGGAUGGCACACUUAUUGAUCUCUGCGGCGCAACCU -------(((..((((((...((((((...((.((-....)).))........(((((...))))).)))))).(.....)...)))))).))).((.....)). ( -17.10, z-score = 0.48, R) >droSim1.chr3R 19502438 92 - 27517382 -------CAGUUAAUUAAAAUUCCAACUUUCCAAC------UAGAUCUAUGACGAAUGCAACAUUCUGCAGGAUGGCACACUUAUUGAUCUCUGCGGCGCAACCU -------.......................((...------.(((((.((((.(..(((..(((((....))))))))..))))).)))))....))........ ( -12.30, z-score = 0.84, R) >droSec1.super_0 20028815 92 - 21120651 -------CAGUUGAUUAAAAUUCCAACUUUUCAAC------UAGAUCUAUGACGAAUGCAACAUUCUGCAGGAUGGCACACUUAUUGAUCUCUGCGGCGCAACCU -------.((((((................)))))------)(((((.((((.(..(((..(((((....))))))))..))))).)))))....((.....)). ( -15.89, z-score = -0.11, R) >droYak2.chr3R 20578257 99 - 28832112 ------GCAGUUGAUUAAAAUUCCUGCUUUCCUUUGCCUACUAGAUCUAUGACGAAUGCAACAUACUGCAGGAUGGCACACUUAUUGAUCUCUGCGGCGCAACAU ------((((..((((((......((((.(((.............((......)).((((......))))))).))))......)))))).)))).......... ( -21.40, z-score = -0.74, R) >droEre2.scaffold_4820 2097607 91 + 10470090 -------CAGUUGAUU-AAAUUCCUACUUUGCCAC------UAGAUCUAUGACGAAUGCAACAUCCUGCAGGAUGGCACACUUAUUGAUCUCUGCGGCGCAACAU -------..((((...-............((((.(------.(((((.((((.(..(((..(((((....))))))))..))))).)))))..).)))))))).. ( -22.06, z-score = -1.80, R) >droAna3.scaffold_13340 3842209 96 + 23697760 ------GAAUUUCAUUAAUCUUGAACGUAUCUUCCU--CU-CAGAUCUAUGAUGAAUGCAACAUCUUGCAGGAUGGCACCCUAAUUGAUCUCUGCGGCGCAACUU ------....((((.......))))........((.--(.-.(((((.((...(..(((..(((((....))))))))..)..)).)))))..).))........ ( -14.00, z-score = 1.28, R) >dp4.chr2 12930922 95 - 30794189 -----UAUCCCUGAUUAAUCUC---UGUGUCUGCCC--UUGUAGGUCUAUGACGAAUGCAACAUCCUGCAGGAUGGCACACUUAUUGAUCUCUGCGGUGCCACGC -----.......(((((((...---((((((....(--(((((((....((.......))....))))))))..))))))...)))))))...(((......))) ( -25.40, z-score = -1.00, R) >droPer1.super_0 4297802 93 + 11822988 -----UAUCCCUGAUUAA--UC---UGUGUCUGCCC--UUGUAGGUCUAUGACGAAUGCAACAUCCUGCAGGAUGGCACACUUAUUGAUCUCUGCGGUGCCACGC -----.......((((((--(.---.((((..((((--(((((((....((.......))....))))))))..)))))))..)))))))...(((......))) ( -26.90, z-score = -1.55, R) >droWil1.scaffold_181089 5891760 95 + 12369635 AGGAAAGAGAUUUUCUAAUGUU-AAUAUAUUA---------CAGAUCUACGAUGAAUGCAACAUCCUGCAGGAUGGCACACUUAUUGAUCUCUGUGGAGCAACGC ((((((....))))))......-......(((---------((((....((((((.((...(((((....)))))...)).))))))...)))))))........ ( -20.50, z-score = -0.85, R) >droVir3.scaffold_13047 865473 95 + 19223366 ------AUUAUAUUAUUAUAUU-CACAUAUUAUGUU---UGCAGAUCUACGAUGAAUGCAACAUCUUGCAGGAUGGCACACUCAUUGAUCUCUGCGGCGCAACGU ------................-.........((((---((((((....((((((.((...(((((....)))))...)).))))))...))))))).))).... ( -20.90, z-score = -0.90, R) >droMoj3.scaffold_6540 13379630 88 - 34148556 --------------CUAAUUUA-UAUGCUCUCAUUU--GCCCAGAUCUAUGAUGAAUGCAACAUAUUGCAGGAUGGCACACUCAUUGAUCUCUGCGGCGCAACGC --------------........-...........((--(((((((((.((((((..(((((....)))))......))...)))).)))))....)).))))... ( -15.90, z-score = 1.11, R) >droGri2.scaffold_15074 6343799 88 - 7742996 --------------CUAAUAAA-UAUAUUUUGUUUC--ACACAGAUCUAUGAUGAGUGUAAUAUCUUGCAGGAUGGCACACUCAUUGAUCUCUGCGGCGCAACGC --------------........-......((((.((--.((.(((((...(((((((((..(((((....)))))..)))))))))))))).)).)).))))... ( -26.20, z-score = -3.18, R) >consensus _______CAGUUGAUUAAUAUU_CAUCUUUUCAAC____U_UAGAUCUAUGACGAAUGCAACAUCCUGCAGGAUGGCACACUUAUUGAUCUCUGCGGCGCAACGU ..........................................(((((.((((.(..(((..(((((....))))))))..))))).))))).............. (-11.89 = -11.48 + -0.41)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:39:00 2011