| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,685,160 – 19,685,280 |
| Length | 120 |
| Max. P | 0.987526 |

| Location | 19,685,160 – 19,685,280 |
|---|---|
| Length | 120 |
| Sequences | 10 |
| Columns | 131 |
| Reading direction | forward |
| Mean pairwise identity | 83.24 |
| Shannon entropy | 0.35334 |
| G+C content | 0.44910 |
| Mean single sequence MFE | -30.28 |
| Consensus MFE | -24.71 |
| Energy contribution | -24.45 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.987526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
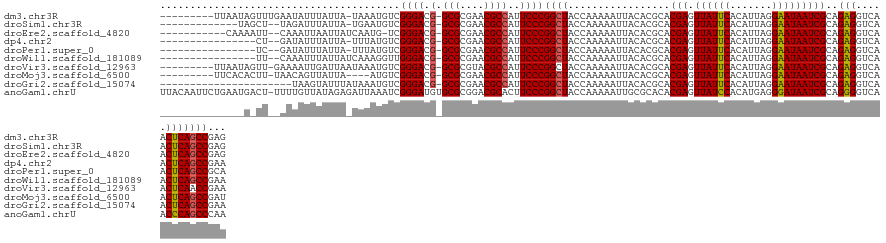
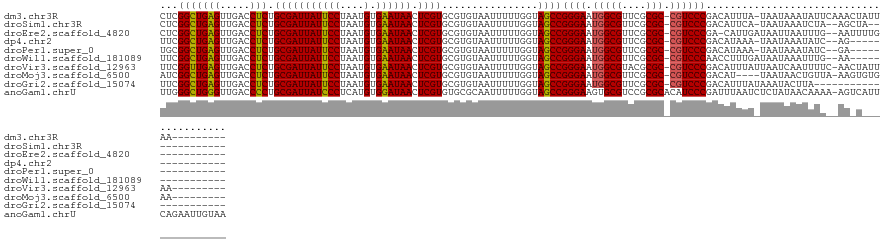
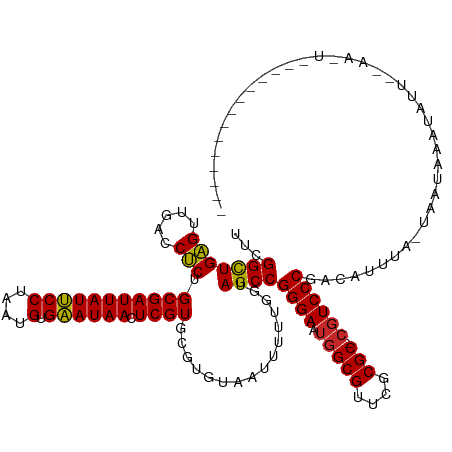
>dm3.chr3R 19685160 120 + 27905053 ---------UUAAUAGUUUGAAUAUUUAUUA-UAAAUGUCGGGACG-GCGCGAACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACAUUAGGAAUAAUCGCAGAGGUCAACUCAGCCGAG ---------.((((((.........))))))-.....(((((((.(-(((....))))..)))))))...............((.(((.((((((.......)))))))))..(((.....))).)).... ( -28.50, z-score = -1.72, R) >droSim1.chr3R 19499823 114 + 27517382 -------------UAGCU--UAGAUUUAUUA-UGAAUGUCGGGACG-GCGCGAACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACAUUAGGAAUAAUCGCAGAGGUCAACUCAGCCGAG -------------.....--...........-((...(((((((.(-(((....))))..)))))))...))..........((.(((.((((((.......)))))))))..(((.....))).)).... ( -28.50, z-score = -1.53, R) >droEre2.scaffold_4820 2094972 116 - 10470090 -----------CAAAAUU--CAAAUUAAUUAUCAAUG-UCGGGACG-GCGCGAACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACAUUAGGAAUAAUCGCAGAGGUCAACUCAGCCGAG -----------.......--...........((...(-((((((.(-(((....))))..)))))))...............((.(((.((((((.......)))))))))..(((.....))).)).)). ( -28.70, z-score = -2.65, R) >dp4.chr2 12924954 111 + 30794189 ----------------CU--GAUAUUUAUUA-UUUAUGUCGGGACG-GCGCGAACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACAUUAGGAAUAAUCGCAGAGGUCAACUCAGCCGAA ----------------..--(((((......-...)))))((((.(-(((....))))..))))((((.................(((.((((((.......)))))))))..(((.....)))))))... ( -29.60, z-score = -2.22, R) >droPer1.super_0 4293616 111 - 11822988 ----------------UC--GAUAUUUAUUA-UUUAUGUCGGGACG-GCGCGAACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACAUUAGGAAUAAUCGCAGAGGUCAACUCAGCCGCA ----------------..--(((((......-...)))))((((.(-(((....))))..))))((((.................(((.((((((.......)))))))))..(((.....)))))))... ( -29.90, z-score = -2.26, R) >droWil1.scaffold_181089 5881056 112 - 12369635 ----------------UU--CAAAUUUAUUAUCAAAGGUUGGGACG-GCGCGAACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACAUUAGGAAUAAUCGCAGAGGUCAACUCAGCCGAA ----------------..--...........((...((((((((.(-(((....))))..))))))))..............((.(((.((((((.......)))))))))..(((.....))).)).)). ( -28.10, z-score = -2.12, R) >droVir3.scaffold_12963 1581635 120 + 20206255 ---------UUAAUAGUU-GAAAAUUGAUUAAUAAAUGUCGGGACG-GCGCGUACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACAUUAGGAAUAAUCGCAGAGGUCAACUCAACCGAA ---------......(((-((...((((((.......(((((((.(-(((....))))..)))))))..................(((.((((((.......)))))))))....)))))).))))).... ( -31.00, z-score = -2.35, R) >droMoj3.scaffold_6500 28482783 116 + 32352404 ---------UUCACACUU-UAACAGUUAUUA----AUGUCGGGACG-GCGCGAACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACAUUAGGAAUAAUCGCAGAGGUCAACUCAGCCGAU ---------.........-............----..(((((((.(-(((....))))..))))((((.................(((.((((((.......)))))))))..(((.....)))))))))) ( -28.50, z-score = -2.23, R) >droGri2.scaffold_15074 1401220 108 - 7742996 ----------------------UAAGUAUUUAUAAAUGUCGGGACG-GCGCGAACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACAUUAGGAAUAAUCGCAGAGGUCAACUCAGCCGAA ----------------------...(((.........(((((((.(-(((....))))..)))))))..........)))..((.(((.((((((.......)))))))))..(((.....))).)).... ( -29.51, z-score = -2.53, R) >anoGam1.chrU 49124114 130 - 59568033 UUACAAUUCUGAAUGACU-UUUUGUUAUAGAGAUUAAAUCGGGAUGUGCGCGGACGCACUUCCCGGCUACCAAAAAUUGCGCACACGAGUUAUCCACAUGAGGGAUAAUCGCAGGGGUCAACCCAGCCCAA .......(((..(((((.-....)))))..))).......((((.(((((....))))).))))((((.........((((.......(((((((.......)))))))))))(((.....)))))))... ( -40.51, z-score = -2.52, R) >consensus ______________A_UU__AAUAUUUAUUA_UAAAUGUCGGGACG_GCGCGAACGCCAUUCCCGGCUACCAAAAAUUACACGCACGAGUUAUUCACAUUAGGAAUAAUCGCAGAGGUCAACUCAGCCGAA ........................................((((...(((....)))...))))((((.................(((.((((((.......)))))))))..(((.....)))))))... (-24.71 = -24.45 + -0.26)
| Location | 19,685,160 – 19,685,280 |
|---|---|
| Length | 120 |
| Sequences | 10 |
| Columns | 131 |
| Reading direction | reverse |
| Mean pairwise identity | 83.24 |
| Shannon entropy | 0.35334 |
| G+C content | 0.44910 |
| Mean single sequence MFE | -32.34 |
| Consensus MFE | -27.52 |
| Energy contribution | -27.07 |
| Covariance contribution | -0.45 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.905682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
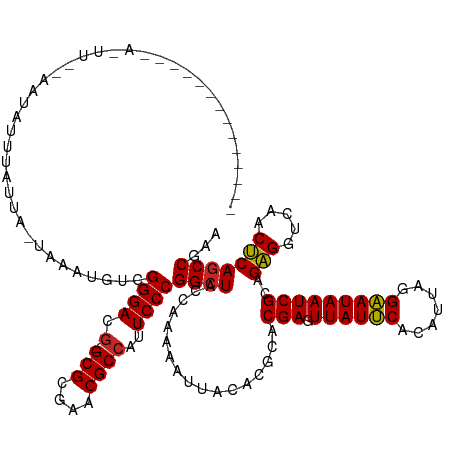
>dm3.chr3R 19685160 120 - 27905053 CUCGGCUGAGUUGACCUCUGCGAUUAUUCCUAAUGUGAAUAACUCGUGCGUGUAAUUUUUGGUAGCCGGGAAUGGCGUUCGCGC-CGUCCCGACAUUUA-UAAUAAAUAUUCAAACUAUUAA--------- ...(((((((.....))).(((((((((((....).)))))).))))................))))((((.(((((....)))-))))))........-......................--------- ( -30.90, z-score = -1.28, R) >droSim1.chr3R 19499823 114 - 27517382 CUCGGCUGAGUUGACCUCUGCGAUUAUUCCUAAUGUGAAUAACUCGUGCGUGUAAUUUUUGGUAGCCGGGAAUGGCGUUCGCGC-CGUCCCGACAUUCA-UAAUAAAUCUA--AGCUA------------- ...(((((((.....))).(((((((((((....).)))))).))))................))))((((.(((((....)))-))))))........-...........--.....------------- ( -30.90, z-score = -1.03, R) >droEre2.scaffold_4820 2094972 116 + 10470090 CUCGGCUGAGUUGACCUCUGCGAUUAUUCCUAAUGUGAAUAACUCGUGCGUGUAAUUUUUGGUAGCCGGGAAUGGCGUUCGCGC-CGUCCCGA-CAUUGAUAAUUAAUUUG--AAUUUUG----------- .(((((.(((.....))).(((((((((((....).)))))).))))))...(((((.(..((.(.(((((.(((((....)))-))))))).-)))..).)))))...))--)......----------- ( -31.90, z-score = -1.52, R) >dp4.chr2 12924954 111 - 30794189 UUCGGCUGAGUUGACCUCUGCGAUUAUUCCUAAUGUGAAUAACUCGUGCGUGUAAUUUUUGGUAGCCGGGAAUGGCGUUCGCGC-CGUCCCGACAUAAA-UAAUAAAUAUC--AG---------------- ...(((((((.....))).(((((((((((....).)))))).))))................))))((((.(((((....)))-))))))........-...........--..---------------- ( -30.90, z-score = -1.39, R) >droPer1.super_0 4293616 111 + 11822988 UGCGGCUGAGUUGACCUCUGCGAUUAUUCCUAAUGUGAAUAACUCGUGCGUGUAAUUUUUGGUAGCCGGGAAUGGCGUUCGCGC-CGUCCCGACAUAAA-UAAUAAAUAUC--GA---------------- .((.((..((...(((.(.(((((((((((....).)))))).))))).).))....))..)).))(((((.(((((....)))-))))))).......-...........--..---------------- ( -31.30, z-score = -0.95, R) >droWil1.scaffold_181089 5881056 112 + 12369635 UUCGGCUGAGUUGACCUCUGCGAUUAUUCCUAAUGUGAAUAACUCGUGCGUGUAAUUUUUGGUAGCCGGGAAUGGCGUUCGCGC-CGUCCCAACCUUUGAUAAUAAAUUUG--AA---------------- ...(((((((.....))).(((((((((((....).)))))).))))................))))((((.(((((....)))-))))))....................--..---------------- ( -30.90, z-score = -1.55, R) >droVir3.scaffold_12963 1581635 120 - 20206255 UUCGGUUGAGUUGACCUCUGCGAUUAUUCCUAAUGUGAAUAACUCGUGCGUGUAAUUUUUGGUAGCCGGGAAUGGCGUACGCGC-CGUCCCGACAUUUAUUAAUCAAUUUUC-AACUAUUAA--------- ...(((((((((((.....(((((((((((....).)))))).))))...........(((((((.(((((.(((((....)))-)))))))....))))))))))))..))-)))).....--------- ( -34.50, z-score = -2.30, R) >droMoj3.scaffold_6500 28482783 116 - 32352404 AUCGGCUGAGUUGACCUCUGCGAUUAUUCCUAAUGUGAAUAACUCGUGCGUGUAAUUUUUGGUAGCCGGGAAUGGCGUUCGCGC-CGUCCCGACAU----UAAUAACUGUUA-AAGUGUGAA--------- .(((((((((.....))).(((((((((((....).)))))).))))................))))((((.(((((....)))-))))))))..(----((...(((....-.))).))).--------- ( -32.10, z-score = -1.09, R) >droGri2.scaffold_15074 1401220 108 + 7742996 UUCGGCUGAGUUGACCUCUGCGAUUAUUCCUAAUGUGAAUAACUCGUGCGUGUAAUUUUUGGUAGCCGGGAAUGGCGUUCGCGC-CGUCCCGACAUUUAUAAAUACUUA---------------------- ...(((((((.....))).(((((((((((....).)))))).))))................))))((((.(((((....)))-))))))..................---------------------- ( -30.90, z-score = -1.46, R) >anoGam1.chrU 49124114 130 + 59568033 UUGGGCUGGGUUGACCCCUGCGAUUAUCCCUCAUGUGGAUAACUCGUGUGCGCAAUUUUUGGUAGCCGGGAAGUGCGUCCGCGCACAUCCCGAUUUAAUCUCUAUAACAAAA-AGUCAUUCAGAAUUGUAA ..(.((.(((....)))..(((((((((((....).)))))).)))))).)((((((((..((....((((.(((((....))))).))))(((((...............)-))))))..)))))))).. ( -39.06, z-score = -1.40, R) >consensus UUCGGCUGAGUUGACCUCUGCGAUUAUUCCUAAUGUGAAUAACUCGUGCGUGUAAUUUUUGGUAGCCGGGAAUGGCGUUCGCGC_CGUCCCGACAUUUA_UAAUAAAUAUU__AA_U______________ ...(((((((.....))).(((((((((((....).)))))).))))................))))((((...(((....)))...))))........................................ (-27.52 = -27.07 + -0.45)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:38:59 2011