| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,683,934 – 19,684,016 |
| Length | 82 |
| Max. P | 0.877767 |

| Location | 19,683,934 – 19,684,016 |
|---|---|
| Length | 82 |
| Sequences | 3 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 56.15 |
| Shannon entropy | 0.57621 |
| G+C content | 0.51318 |
| Mean single sequence MFE | -18.42 |
| Consensus MFE | -9.75 |
| Energy contribution | -8.20 |
| Covariance contribution | -1.55 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.773765 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
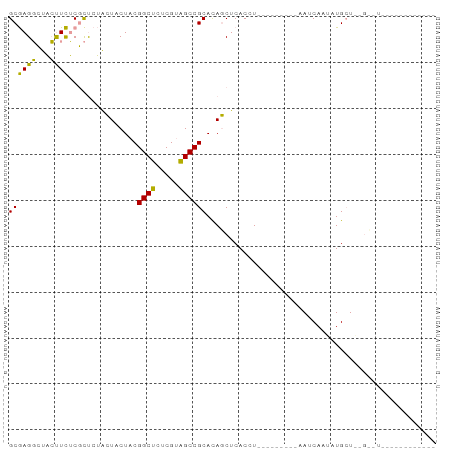
>dm3.chr3R 19683934 82 + 27905053 GCAAGUUCCACUUGCAUCAGGGUUCU-CGGCACUCGCUGCCGCAUACCUCACCUUGAUAUGACAACCAACAUGCUGUGGUUCU---------- (((((.....)))))((((((((...-(((((.....)))))........)))))))).....(((((........)))))..---------- ( -23.90, z-score = -1.95, R) >droSim1.chrU 10347172 84 - 15797150 GCGAGGCUACUUCUCGCUCUACUACUACGGCUCUCGUAGCCGCACAGCUCACCU---------AAUCAAUAUGAUAAGCAUUCCUAGGGUUCU ((((((.....))))))..........(((((.....))))).........(((---------(......(((.....)))...))))..... ( -16.90, z-score = 0.01, R) >droSec1.super_86 73009 66 + 128402 GCGAGGCUACUUCUCGCUCUACUACUACGGCUCUCGUAGCCGCACAGCUCACUU---------AAUCAAUAUGCU------------------ ((((((.....))))))..........(((((.....)))))...(((......---------.........)))------------------ ( -14.46, z-score = -1.27, R) >consensus GCGAGGCUACUUCUCGCUCUACUACUACGGCUCUCGUAGCCGCACAGCUCACCU_________AAUCAAUAUGCU__G__U____________ ((((((...))))...............((((.....)))))).................................................. ( -9.75 = -8.20 + -1.55)


| Location | 19,683,934 – 19,684,016 |
|---|---|
| Length | 82 |
| Sequences | 3 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 56.15 |
| Shannon entropy | 0.57621 |
| G+C content | 0.51318 |
| Mean single sequence MFE | -22.60 |
| Consensus MFE | -13.48 |
| Energy contribution | -13.60 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.877767 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
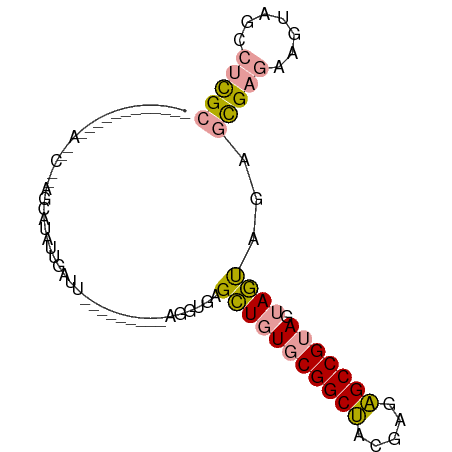
>dm3.chr3R 19683934 82 - 27905053 ----------AGAACCACAGCAUGUUGGUUGUCAUAUCAAGGUGAGGUAUGCGGCAGCGAGUGCCG-AGAACCCUGAUGCAAGUGGAACUUGC ----------....((((.((((...((((.(((((((.......)))))..((((.....)))))-).))))...))))..))))....... ( -24.10, z-score = -0.58, R) >droSim1.chrU 10347172 84 + 15797150 AGAACCCUAGGAAUGCUUAUCAUAUUGAUU---------AGGUGAGCUGUGCGGCUACGAGAGCCGUAGUAGUAGAGCGAGAAGUAGCCUCGC ......(((.....(((((((.((.....)---------))))))))..(((((((.....)))))))....))).(((((.......))))) ( -24.00, z-score = -0.99, R) >droSec1.super_86 73009 66 - 128402 ------------------AGCAUAUUGAUU---------AAGUGAGCUGUGCGGCUACGAGAGCCGUAGUAGUAGAGCGAGAAGUAGCCUCGC ------------------(((.((((....---------.)))).))).(((((((.....)))))))........(((((.......))))) ( -19.70, z-score = -1.45, R) >consensus ____________A__C__AGCAUAUUGAUU_________AGGUGAGCUGUGCGGCUACGAGAGCCGUAGUAGUAGAGCGAGAAGUAGCCUCGC .............................................(((((((((((.....))))))).))))...(((((.......))))) (-13.48 = -13.60 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:38:57 2011