
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,672,904 – 19,673,016 |
| Length | 112 |
| Max. P | 0.647284 |

| Location | 19,672,904 – 19,673,016 |
|---|---|
| Length | 112 |
| Sequences | 13 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 72.29 |
| Shannon entropy | 0.63179 |
| G+C content | 0.50562 |
| Mean single sequence MFE | -31.11 |
| Consensus MFE | -16.19 |
| Energy contribution | -15.79 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.40 |
| Mean z-score | -0.70 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.647284 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 19672904 112 + 27905053 ---CCUAAAUGUGCUCUCUCUUAUUUCCCAAGACCCAACCUACACUACGACGGAGCUGGACCUGAUGCCUACUUUUGGGUGGGCAAUGGCAGCGAACCAAACAUCAUGGGCAUAA ---......(((((((..((((.......))))..................((.((((..((...((((((((....))))))))..))))))...)).........))))))). ( -29.30, z-score = -0.71, R) >droSim1.chr3R 19495209 109 + 27517382 ---ACUAAAUAUACUUCCUUGUUUUCC---AGACCCAACCUGCACUAUGACGGAGCUGGACCUGAUGCCUACUUUUGGGUGGGCAAUGGCAGCGAACCAAACAUCAUGGGCAUAA ---........................---..........(((.((((((.((.((((..((...((((((((....))))))))..))))))...)).....)))))))))... ( -27.90, z-score = -0.48, R) >droSec1.super_0 20021612 109 + 21120651 ---ACUAAAUGUGCUCUCUUCUUUUUC---AGACCCAACCUGCACUACGACGGAGCUGGACCUGAUGCCUACUUUUGGGUGGGCAAUGGCAGCGAACCAAACAUCAUGGGCAUAA ---......(((((((.........((---((..(((.((((........))).).)))..))))((((((((....)))))))).(((.......)))........))))))). ( -30.10, z-score = -1.00, R) >droEre2.scaffold_4820 2090123 109 - 10470090 ---CCUGAGUGCUCGUUCUUGCUUUCC---AGACCCAACCUGCACUACGACGGAGCUGGACCUGAUGCCUACUUCUGGGUGGGCAAUGGCAGCGAACCAAACAUCAUGGGCAUAA ---.....(((((((((((((((.(((---((..((...............))..))))).....((((((((....))))))))..))))).))))..........)))))).. ( -33.26, z-score = -0.46, R) >droYak2.chr3R 20571125 109 + 28832112 ---CCCAAGUACACGCUCUUGUUUUUU---AGACCCAACCUGCACUACGAUGGAGCUGGACCUGAUGCCUACUUCUGGGUGGGCAAUGGCAGCGAACCCAACAUCAUGGGCAUAA ---..((((........))))......---..........(((.(((.((((..((((..((...((((((((....))))))))..))))))(....)..)))).))))))... ( -28.70, z-score = 0.32, R) >droAna3.scaffold_13340 3834164 109 - 23697760 ---AUUAACGGCUCUUAUUUAUUUUAC---AGUCCCAAUCUGCAUUACGAUGGCGCUGGACCGGAUGCUUACUUUUGGGUGGGCAACGGCAGUGAACCGAACAUCAUGGGCGUUA ---..(((((.(((............(---((.......)))......((((.(((((..(((..((((((((....)))))))).)))))))).......))))..)))))))) ( -29.10, z-score = -0.43, R) >dp4.chr2 12917674 111 + 30794189 ---UAUAACCACUUUCCCCUCGCCCCUCC-AGACCGAAUCUGCACUAUGAUGGCGCCGGACCCGACGCCUACUUCUGGGUGGGCAACGGCAGCGAACCCAACAUCAUGGGCAUAA ---..................((((...(-(((.....))))....(((((((((((((......)(((((((....)))))))..)))).))(....)..)))))))))).... ( -32.80, z-score = -0.81, R) >droPer1.super_0 4282518 111 - 11822988 ---AAUAAACACUCUGCCCUCGCCCCUCC-AGACCGAAUCUGCACUAUGAUGGCGCCGGACCCGACGCCUACUUCUGGGUGGGCAACGGCAGCGAACCCAACAUCAUGGGCAUAA ---........(.(((((...((((.(((-(((....(((........)))((((.((....)).))))....)))))).))))...))))).)..((((......))))..... ( -40.90, z-score = -3.15, R) >droWil1.scaffold_181089 5871689 104 - 12369635 ----------UUGAUAAAUUUUUGCUUAC-AGACCCAAUUUGCAUUAUGAUGGAGCCGGACCUGAUGCCUACUUCUGGGUGGGCAAUGGCAGUGAGCCAAACAUUAUGGGAAUUA ----------((.((((..(((.((((((-....(((.((........))))).((((.......((((((((....)))))))).)))).)))))).)))..)))).))..... ( -25.60, z-score = -0.07, R) >droVir3.scaffold_13047 854464 114 - 19223366 GCAAAU-UGUAUCUACAAUAUAUUUAUGACAGUCCCAACCUGCACUAUGACGGCGCUGGACCCGAUGCCUAUUUCUGGGUGGGCAAUGGAAGCGAGCCGAAUCUCAUGGGCAUUA ....((-(((....))))).....................(((.(((((((((((((....(((.((((((((....)))))))).))).)))..))))....)))))))))... ( -32.20, z-score = -0.44, R) >droMoj3.scaffold_6540 21078801 112 - 34148556 GUAUAU-UCUCGGUAUCGUUCAUUUUU--CAGUCCCAAUCUGCACUAUGAUGGCGCCGGACCCGAUGCCUAUUUCUGGGUGGGCAAUGGCAGCGAACCGAAUAUUAUGGGCAUUA ......-((.((((..(((.(((....--(((.......)))....))))))..))))))...(((((((((...(.(((..((.......))..))).).....))))))))). ( -29.70, z-score = 0.55, R) >droGri2.scaffold_15074 6329022 113 + 7742996 AUAAAUAUAUAUGUA-ACUAUAAUUGUG-UAGACCCAAUCUGCAUUACGAUGGCGCUGGACCCGAUGCCUAUUUUUGGGUGGGCAAUGGCAGCGAGCCAAAUAUCAUGGGCAUUA ...............-.((((....(((-((((.....)))))))....))))(((((...(((.((((((((....)))))))).)))))))).(((..........))).... ( -32.60, z-score = -0.68, R) >apiMel3.Group1 13419371 88 + 25854376 --------------------------UAC-AUACCGAAUCUUCAUUACGAUGGUGCCGGGCCGGACGCGUAUUUCUGGGUUGGGAACGGUUCAGAGCCGACCACGUUCGGCAUCA --------------------------...-...(((.(((.((.....)).)))..)))((((((((.((..(((((..(((....)))..)))))...))..)))))))).... ( -32.30, z-score = -1.70, R) >consensus ___AAUAAAUAUGCUCACUUCUUUUUU___AGACCCAAUCUGCACUACGAUGGAGCUGGACCUGAUGCCUACUUCUGGGUGGGCAAUGGCAGCGAACCAAACAUCAUGGGCAUAA .................................((((.((.((.((.....)).)).)).......(((((((....)))))))..(((.......))).......))))..... (-16.19 = -15.79 + -0.39)

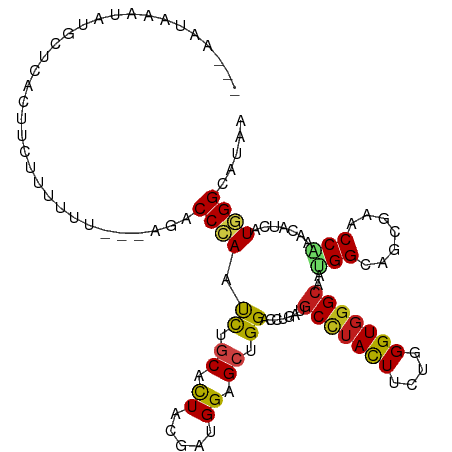

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:38:55 2011