
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,652,727 – 19,652,826 |
| Length | 99 |
| Max. P | 0.766644 |

| Location | 19,652,727 – 19,652,826 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 89.92 |
| Shannon entropy | 0.20148 |
| G+C content | 0.34992 |
| Mean single sequence MFE | -16.80 |
| Consensus MFE | -15.85 |
| Energy contribution | -15.84 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.766644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 19652727 99 + 27905053 ------------UUUCCAAUUCUCUU-----GGCUUUGACACAAGCAACAAUUUACUCGGUUUCAGGUUAAAGUCAUAUCGUUAGCAUUAACGCUUUUUGAUUUUUAUUGAGUCAG ------------.........(((.(-----(((((((((..((((.............))))...)))))))))).((((..(((......)))...)))).......))).... ( -17.32, z-score = -1.40, R) >droSim1.chr3R 19484205 99 + 27517382 ------------CUCCCAAUUCUCUU-----GGCUUUGACACAAGCAACAAUUUACUCGGUUUCAGGUUAAAGUCAUAUCGUUAGCAUUAACGCUUUUUGAUUUUUAUUGAGUCAG ------------.........(((.(-----(((((((((..((((.............))))...)))))))))).((((..(((......)))...)))).......))).... ( -17.32, z-score = -1.20, R) >droSec1.super_0 20010657 99 + 21120651 ------------AUCCCAAUUCUCUU-----GGCUUUGACACAAGCAACAAUUUACUCGGUUUCAGGUUAAAGUCAUAUCGUUAGCAUUAACGCUUUUUGAUUUUUAUUGAGUCAG ------------.........(((.(-----(((((((((..((((.............))))...)))))))))).((((..(((......)))...)))).......))).... ( -17.32, z-score = -1.32, R) >droYak2.chr3R 20559744 104 + 28832112 ------------UUUCCCAUUCUUUUUUCUCGGCUUUGACACAAGCAACAAUUUACUCGGUUUCAGGUUAAAGUCAUAUCGUUAGCAUUAACGCUUUUUGAUUUUUAUUGAGUCAG ------------................((((((((((((..((((.............))))...)))))))))..((((..(((......)))...)))).......))).... ( -17.42, z-score = -1.70, R) >droEre2.scaffold_4820 2078995 101 - 10470090 ------------UUUCCUAUUCUUUUUU---GGCUUUGACACAAGCAACAAUUUACUCGGUUUCAGGUUAAAGUCAUAUCGUUAGCAUUAACGCUUUUUGAUUUUUAUUGAGUCAG ------------...............(---(((((((((..((((.............))))...))))))))))...(((((....))))).....((((((.....)))))). ( -16.52, z-score = -1.23, R) >droAna3.scaffold_13340 3821309 101 - 23697760 ------------AUCCCCACCCACUUUU---GGCUUUGACACAAGCAACAAUUUACUCGGUUUCAGGUUAAAGUCAUAUCGUUAGCAUUAACGCUUUUUGAUUUUUAUUGAGUCAG ------------...............(---(((((((((..((((.............))))...))))))))))...(((((....))))).....((((((.....)))))). ( -16.52, z-score = -0.98, R) >dp4.chr2 12906414 101 + 30794189 ------------UCCCCCUUUUUUUUUU---GGCUUUGACACAAGCAACAAUUUACUCGGUUUCAGGUUAAAGUCAUAUCGUUAGCAUUAACGCUUUUUGAUUUUUAUUGAGUCAG ------------...............(---(((((((((..((((.............))))...))))))))))...(((((....))))).....((((((.....)))))). ( -16.52, z-score = -1.07, R) >droPer1.super_0 4271181 102 - 11822988 -----------UCCCCCUUUUUUUUUUU---GGCUUUGACACAAGCAACAAUUUACUCGGUUUCAGGUUAAAGUCAUAUCGUUAGCAUUAACGCUUUUUGAUUUUUAUUGAGUCAG -----------................(---(((((((((..((((.............))))...))))))))))...(((((....))))).....((((((.....)))))). ( -16.52, z-score = -1.02, R) >droVir3.scaffold_13047 839154 96 - 19223366 -----------------ACCCCCUUUUU---GGCUUUGACACAAGCAACAAUUUACUCGGUUUCAGGUUAAAGUCAUAUCGUUAGCAUUAACGCUUUUUGAUUUUUAUUGAGUCAG -----------------..........(---(((((((((..((((.............))))...))))))))))...(((((....))))).....((((((.....)))))). ( -16.52, z-score = -1.01, R) >droMoj3.scaffold_6540 21065069 98 - 34148556 ---------------ACCCCCUUUUUUU---GGCUUUGACACAAGCAACAAUUUACUCGGUUUCAGGUUAAAGUCAUAUCGUUAGCAUUAACGCUUUUUGAUUUUUAUUGAGUCAG ---------------............(---(((((((((..((((.............))))...))))))))))...(((((....))))).....((((((.....)))))). ( -16.52, z-score = -1.04, R) >droGri2.scaffold_15074 6315796 113 + 7742996 ACCCCCUUUUCUUUUUUUUUUUUUGUUU---GGCUUUGACACAAGCAACAAUUUACUCGGUUUCAGGUUAAAGUCAUAUCGUUAGCAUUAACGCUUUUUGAUUUUUAUUGAGUCAG ...........................(---(((((((((..((((.............))))...))))))))))...(((((....))))).....((((((.....)))))). ( -16.52, z-score = -0.63, R) >droWil1.scaffold_181089 5856992 97 - 12369635 ----------------UCCCCUUUUUUU---GGCUUUGACACAAGCAACAAUUUACUCGGUUUCAGGUUAAAGUCAUAUCGUUAGCAUUAACGCUUUUUGAUUUUUAUUGAGUCAG ----------------...........(---(((((((((..((((.............))))...))))))))))...(((((....))))).....((((((.....)))))). ( -16.52, z-score = -1.09, R) >consensus ____________UUCCCCAUUCUUUUUU___GGCUUUGACACAAGCAACAAUUUACUCGGUUUCAGGUUAAAGUCAUAUCGUUAGCAUUAACGCUUUUUGAUUUUUAUUGAGUCAG ...............................(((((((((..((((.............))))...)))))))))....(((((....))))).....((((((.....)))))). (-15.85 = -15.84 + -0.00)

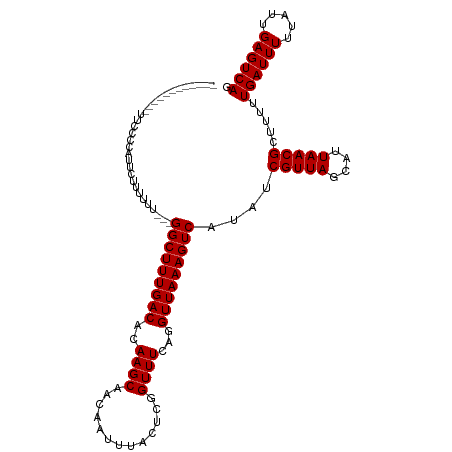
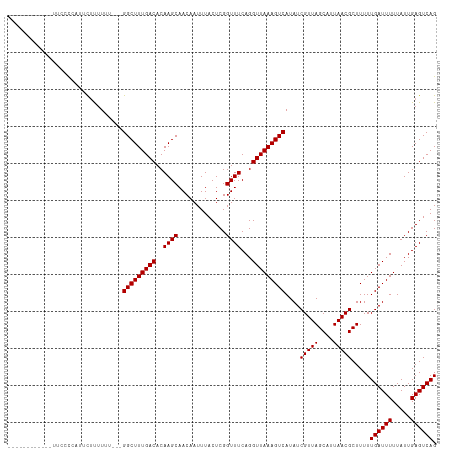
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:38:53 2011