
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,646,539 – 19,646,637 |
| Length | 98 |
| Max. P | 0.836483 |

| Location | 19,646,539 – 19,646,637 |
|---|---|
| Length | 98 |
| Sequences | 9 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 66.76 |
| Shannon entropy | 0.62436 |
| G+C content | 0.30021 |
| Mean single sequence MFE | -13.86 |
| Consensus MFE | -7.00 |
| Energy contribution | -7.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.836483 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
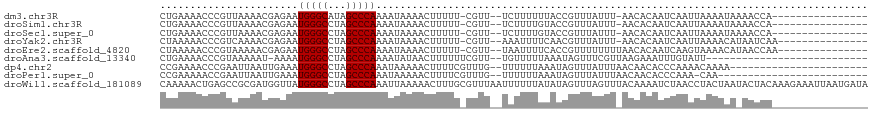
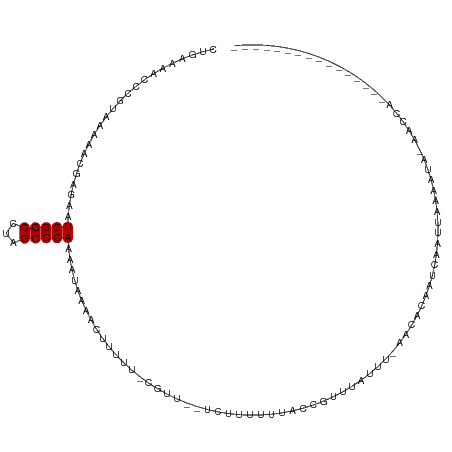
>dm3.chr3R 19646539 98 + 27905053 CUGAAAACCCGUUAAAACGAGAAUGGGCAUAGCCCAAAAUAAAACUUUUU-CGUU--UCUUUUUUACCGUUUAUUU-AACACAAUCAAUUAAAAUAAAACCA---------------- .(((..........((((((((((((((...)))))..........))))-))))--)..........(((.....-)))....)))...............---------------- ( -13.20, z-score = -1.12, R) >droSim1.chr3R 19478086 98 + 27517382 CUGAAAACCCGUUAAAACGAGAAUGGGCCUAGCCCAAAAUAAAACUUUUU-CGUU--UCUUUUGUACCGUUUAUUU-AACACAAUCAAUUAAAAUAAAACCA---------------- ..........(((((((((....(((((...)))))(((..((((.....-.)))--)..)))....)))...)))-)))......................---------------- ( -13.50, z-score = -0.88, R) >droSec1.super_0 20004561 98 + 21120651 CUGAAAACCCGUUAAAACGAGAAUGGGCCUAGCCCAAAAUAAAACUUUUU-CGUU--UCUUUUGUACCGUUUAUUU-AACACAAUCAAUUAAAAUAAAACCA---------------- ..........(((((((((....(((((...)))))(((..((((.....-.)))--)..)))....)))...)))-)))......................---------------- ( -13.50, z-score = -0.88, R) >droYak2.chr3R 20553479 99 + 28832112 CUAAAAACCCGUCAAAACGAGAAUGGGCCUAGCCCAAAAUAAAACUUUUU-CGUU--AAAUUUUCAACGUUUAUUU-AACACAAUCAAUUAAAACAUAAUCAA--------------- ...............(((((((((((((...)))))..........))))-))))--...........(((.....-))).......................--------------- ( -12.00, z-score = -1.67, R) >droEre2.scaffold_4820 2072838 100 - 10470090 CUAAAAACCCGUAAAAACGAGAAUGGGCCUAGCCCAAAAUAAAACUUUUU-CGUU--UAAUUUUCACCGUUUUUUUUAACACAAUCAAGUAAAACAUAACCAA--------------- ..............((((((((((((((...)))))..........))))-))))--)..........(((((.................)))))........--------------- ( -13.23, z-score = -1.20, R) >droAna3.scaffold_13340 3814109 88 - 23697760 CUGAAAACCCGUAAAAAU-AAAAUGGGCCUAGCCCAAAAUAUAACUUUUUUCGUU--UGUUUUUAAAUAGUUUCGUUAAGAAAUUUGUAUU--------------------------- ..((((.....(((((((-(((.(((((...)))))(((........)))...))--)))))))).....)))).................--------------------------- ( -13.40, z-score = -1.00, R) >dp4.chr2 12898370 93 + 30794189 CCGAAAACCCGAAUUAAUUGAAAUGGGCCUAGCCCAAAUAAAAACUUUUCGUUUG--UUUUUUAAAUAGUUUAUUUAACAACACCCAAAACAAAA----------------------- .((((((..(((.....)))...(((((...))))).........))))))((((--((((.......(((.....))).......)))))))).----------------------- ( -13.74, z-score = -1.22, R) >droPer1.super_0 4263242 90 - 11822988 CCGAAAAACCGAAUUAAUUGAAAUGGGCCUAGCCCAAAUAAAAACUUUUCGUUUG--UUUUUUAAAUAGUUUAUUUAACAACACCCAAA-CAA------------------------- ..........((((((.(((((((((((...))))).....((((.....)))).--..)))))).)))))).................-...------------------------- ( -12.30, z-score = -0.78, R) >droWil1.scaffold_181089 5847581 118 - 12369635 CAAAAACUGAGCCGCGAUGGUUAUGGGCCUAGCCCAAAUUAAAAACUUUGCGUUUAAUUUUUUAUAUAGUUUAGUUUACAAAAUCUAACCUACUAAUACUACAAAGAAAUUAAUGAUA ............(((((.(.((((((((...)))))...)))...).))))).((((((((((...(((((((((................))))).))))..))))))))))..... ( -19.89, z-score = -1.00, R) >consensus CUGAAAACCCGUAAAAACGAGAAUGGGCCUAGCCCAAAAUAAAACUUUUU_CGUU__UCUUUUUUACCGUUUAUUU_AACACAAUCAAUUAAAAUA_AACCA________________ .......................(((((...))))).................................................................................. ( -7.00 = -7.00 + 0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:38:52 2011