
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,632,617 – 19,632,722 |
| Length | 105 |
| Max. P | 0.896905 |

| Location | 19,632,617 – 19,632,722 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 77.87 |
| Shannon entropy | 0.49074 |
| G+C content | 0.51214 |
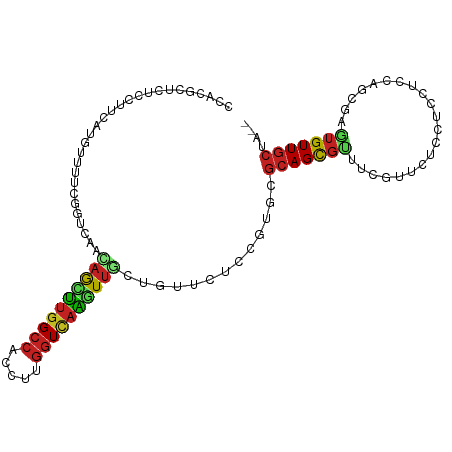
| Mean single sequence MFE | -26.97 |
| Consensus MFE | -12.74 |
| Energy contribution | -12.30 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.65 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.896905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
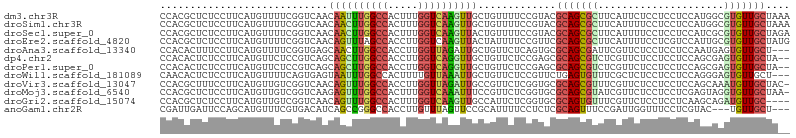
>dm3.chr3R 19632617 105 + 27905053 CCACGCUCUCCUUCAUGUUUUCGGUCAACAAUUUGGCCACUUUGGUCAAGUUGCUGUUUUCCGUACGCAGCGCUUCAUUCUCCUCCUCCAUGGCGUGUUGCUAAA ................((...(((.((.((((((((((.....)))))))))).))....))).))(((((((.((((...........)))).))))))).... ( -29.30, z-score = -3.51, R) >droSim1.chr3R 19464119 105 + 27517382 CCACGCUCUCCUUCAUGUUUUCGGUCAACAACUUGGCCACUUUGGUCAAGUUGCUGUUUUCCGUACGCAGCGCUUCAUUUUCCUCCUCCAUGGCGUGUUGCUAAA ................((...(((.((.((((((((((.....)))))))))).))....))).))(((((((.((((...........)))).))))))).... ( -31.60, z-score = -4.18, R) >droSec1.super_0 19990784 105 + 21120651 CCACGCUCUCCUUCAUGUUUUCGGUCAACAACUUGGCCACUUUGGUCAAGUUACUGUUUUCCGUACGCAGCGCUUCAUUUUCCUCCUCCAUCGCGUGUUGCUAGA ................((...(((..((((((((((((.....))))))))...))))..))).))(((((((.....................))))))).... ( -26.40, z-score = -3.02, R) >droEre2.scaffold_4820 2058884 105 - 10470090 CCACGCUCUCCUUCAUGUUUUCGGUCAACAGUUUAGCCACCUUGGUCAAGUUACUAUUUUCCGUUCGCAGCGCUUCAUUUUCCUCGUCCAUUGCGUGUUGCUAUG .(((((.........(((...(((..((.(((...(((.....)))......))).))..)))...)))(((............))).....)))))........ ( -14.90, z-score = -0.01, R) >droAna3.scaffold_13340 3798935 102 - 23697760 CCACACUUUCCUUCAUGUUUUCGGUGAGCAACUUGGCCACCUUGGUUAGAUUGCUGUUCUCAGUGCGCAGCGAUUCGUUCUCCUCCUCCAAUGAGUGUUGCU--- ..((((......((((......((.((((....(((((.....)))))(((((((((.(.....).))))))))).)))).)).......))))))))....--- ( -25.12, z-score = -0.55, R) >dp4.chr2 12881053 103 + 30794189 CCACACUCUCCUUCAUGUUCUCCGUCAGCAGCUUGGCCACCUUGGUCAGGUUGCUGUUCUCCGAGCGCAGCGUCUCGUUCUCCUCCUCCAGCGAGUGUUGCUA-- ................((((.....(((((((((((((.....)))))))))))))......))))((((((.((((((..........))))))))))))..-- ( -39.70, z-score = -4.93, R) >droPer1.super_0 4247308 103 - 11822988 CCACACUCUCCUUCAUGUUCUCCGUCAGCAGCUUGGCCACCUUGGUCAGGUUGCUGUUCUCCGAGCGCAGCGUCUCGUUCUCCUCCUCCAGCGAGUGUUGCUA-- ................((((.....(((((((((((((.....)))))))))))))......))))((((((.((((((..........))))))))))))..-- ( -39.70, z-score = -4.93, R) >droWil1.scaffold_181089 5830447 102 - 12369635 CAACACUCUCCUUCAUGUUUUCAGUGAGUAAUUUGGCCACUUUUGUUAAAUUGCUGUUCUCCGUUCUGAGUGUUUCGCUCUCCUCCUCCAGGGAGUGUUGCU--- ((((((((.(((..............(((((((((((.......)))))))))))............(((((...))))).........)))))))))))..--- ( -27.40, z-score = -2.87, R) >droVir3.scaffold_13047 814020 104 - 19223366 CCACGCUUUCCUUCAUGUUGUCGGUCAACAGUUUGGCCACCUUGGUUAGAUUGCCGUUCUCGGUGCGCAGCGUUUCGUUCUCCUCCUCCAGCAAAUGUUGCUAC- ....((..........((((.....))))(((((((((.....)))))))))((((....))))))(((((((((.(((..........))))))))))))...- ( -26.50, z-score = -1.19, R) >droMoj3.scaffold_6540 21042085 104 - 34148556 CCACGCUCUCCUUCAUGUUGUCGGUCAAGAGUUUGGCCACUUUGGUCAAAUUUCCGUUCUCGGUGCGCAGCGUAUCGUUCUCCUCCUCGAGUAGGUGUUGCUAA- ....((.(.(((.(((((((.((.....((((((((((.....))))))))))(((....)))..)))))))).(((..........)))).))).)..))...- ( -26.30, z-score = -1.00, R) >droGri2.scaffold_15074 6291666 101 + 7742996 CCACGCUCUCCUUCAUGUUGUCGGUCAACAGUUUGGCCACUUUGGUCAAGUUGCCAUUCUCGGUGCGCAGUGUUUCGUUCUCCUCCUCAAGCAGAUGUUGC---- ....((...((....(((((.....)))))(..((((.((((.....)))).))))..)..)).))((((..(((.(((..........))))))..))))---- ( -22.40, z-score = 0.15, R) >anoGam1.chr2R 47564882 99 + 62725911 CGAUUGAUUCCAGCAUGUUUCGUGACAUCAGCCGGGCCACCUUGUUUAGUUCCGCAUUUUCCUCUCGCAGUUUCCGAUUGGUUUCCUCGUAC---UGUUGCU--- ..........(((((.((..((.((.((((..((((....((.....))....((...........))....))))..)))).))..)).))---)))))..--- ( -14.30, z-score = 2.08, R) >consensus CCACGCUCUCCUUCAUGUUUUCGGUCAACAGCUUGGCCACCUUGGUCAAGUUGCUGUUCUCCGUGCGCAGCGUUUCGUUCUCCUCCUCCAGCGAGUGUUGCUA__ ............................((((((((((.....)))))))))).............(((((((.....................))))))).... (-12.74 = -12.30 + -0.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:38:51 2011