
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,578,244 – 19,578,344 |
| Length | 100 |
| Max. P | 0.584652 |

| Location | 19,578,244 – 19,578,344 |
|---|---|
| Length | 100 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 68.55 |
| Shannon entropy | 0.62648 |
| G+C content | 0.43503 |
| Mean single sequence MFE | -26.10 |
| Consensus MFE | -10.91 |
| Energy contribution | -9.92 |
| Covariance contribution | -0.99 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.584652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
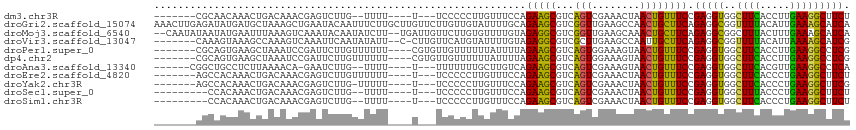
>dm3.chr3R 19578244 100 - 27905053 -------CGCAACAAACUGACAAACGAGUCUUG--UUUU----U---UCCCCCUUGUUUCCAGAAGCGUCAGUCGAAACUAACUGUUUCCGAGGUGGCUUCACCUUGAAGGCUUCU -------.((.....((((((((((((...)))--))).----.---........((((....)))))))))).(((((.....)))))(((((((....)))))))...)).... ( -24.90, z-score = -0.80, R) >droGri2.scaffold_15074 6230509 116 - 7742996 AAACUUGAGAUAUGAUGCUAAAGCUGAAUACAAUUUCUUGCUUGUUCUUGUUGUAUUUUGCAGAAGCGUCGGUUGAAGCCAACUGCUUCAGAGGCGGUUUUACAUUGAAAGCAUCA ............(((((((...((.(((((((((...............))))))))).))((((.((((..(((((((.....))))))).)))).))))........))))))) ( -34.36, z-score = -2.06, R) >droMoj3.scaffold_6540 25952845 112 - 34148556 --CAAUAUAAUAUGAAUUUAAAGUCAAAUACAAUAUCUU--UGAUUGUUCUUGUGUUUUGUAGAGGCGUCGGUUGAAGCAAACUGCUUCAGAGGCGGCUUUACUUUGAAAGCAUCA --.........(((..((((((...((((((((...(..--.....)...)))))))).((((((.((((..(((((((.....))))))).)))).))))))))))))..))).. ( -30.20, z-score = -2.55, R) >droVir3.scaffold_13047 753039 106 + 19223366 -------CAAAGUAAAGCCAAAGUCAAAUUCAAUAUAUU--C-CUUGUUCAUGUAUUUUGUAGAGGCGUCGCUUGAAGCCAAUUGCUUCAGAGGCGGUUUUACAUUAAAAGCAUCG -------....((((((((....................--(-(((...((.......))..)))).(((..(((((((.....))))))).)))))))))))............. ( -25.60, z-score = -2.50, R) >droPer1.super_0 4187431 105 + 11822988 -------CGCAGUGAAGCUAAAUCCGAUUCUUGUUUUUU----CGUGUUGUUUUUUAUUUUAGAAGCGUCAGUGGAAAGUAACUGUUUCCGAGGUGGCUUCACCUUGAAGGCCUCG -------....(((((((((..((.((..(....(((((----((((.(((((((......))))))).)).))))))).....)..)).))..)))))))))............. ( -27.00, z-score = -1.19, R) >dp4.chr2 12822249 105 - 30794189 -------CGCAGUGAAGCUAAAUCCGAUUCUUGUUUUUU----CGUGUUGUUUUUUAUUUUAGAAGCGUCAGUGGAAAGUAACUGUUUCCGAGGUGGCUUCACCUUGAAGGCCUCG -------....(((((((((..((.((..(....(((((----((((.(((((((......))))))).)).))))))).....)..)).))..)))))))))............. ( -27.00, z-score = -1.19, R) >droAna3.scaffold_13340 3742519 100 + 23697760 ------CGGCUGCCUCUUAAAACA-GAAUCUUG--UUUU----U---UUUUUUUGCUUGUCAGAAGCGUCAGUCGAAAGUAACUGUUUCCGAGGUGGCUUCACGUUGAAGGCCUCA ------.(((.(((((...(((((-(.......--....----.---.......((((.....))))......(....)...))))))..)))))..((((.....)))))))... ( -24.30, z-score = -0.03, R) >droEre2.scaffold_4820 2003674 102 + 10470090 -------AGCCACAAACUGACAAACGAGUCUUGUUUUUU----U---UCCCCCUUGUUUCCAGAAGCGUCAGUCGAAACUAACUGUUUCCGAGGUGGCUUCACCCUGAAGGCUUCU -------((((....((((((((((((...))))))...----.---........((((....)))))))))).(((((.....)))))((.((((....)))).))..))))... ( -24.10, z-score = -0.53, R) >droYak2.chr3R 20484632 101 - 28832112 -------AGCCACAAACUGACAAACGAGUCUUG-UUUUU----U---UCCCCCUUGUUUCCAGAAGCGUCAGUCGAAACUAACUGUUUCCGAGGUGGCUUCACCCUGAAGGCUUCG -------((((....((((((((((((...)))-)))..----.---........((((....)))))))))).(((((.....)))))((.((((....)))).))..))))... ( -24.10, z-score = -0.62, R) >droSec1.super_0 19936526 98 - 21120651 ---------CCACAAACUGACAAACGAGUCUUG--UUUU----U---UCCCCCUUGUUUCCAGAAGCGUCAGUCGAAACUAACUGUUUCCGAGGUGGCUUUACCCUGAAGGCUUCU ---------....((((.(((......)))..)--))).----.---..............((((((.(((((((((((.....)))).)))((((....))))))))..)))))) ( -21.60, z-score = -0.26, R) >droSim1.chr3R 19409528 98 - 27517382 ---------CCACAAACUGACAAACGAGUCUUG--UUUU----U---UCCCCCUUGUUUCCAGAAGCGUCAGUCGAAACUAACUGUUUCCGAGGUGGCUUCACCCUGAAGGCUUCU ---------....((((.(((......)))..)--))).----.---..............((((((.(((((((((((.....)))).)))((((....))))))))..)))))) ( -23.90, z-score = -0.84, R) >consensus _______CGCAACAAACUGAAAAACGAGUCUUGUUUUUU____U___UUCCUCUUGUUUCCAGAAGCGUCAGUCGAAACUAACUGUUUCCGAGGUGGCUUCACCUUGAAGGCUUCG ..............................................................(((((...(((........)))))))).(((((..((((.....))))))))). (-10.91 = -9.92 + -0.99)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:38:48 2011