
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,555,902 – 19,555,996 |
| Length | 94 |
| Max. P | 0.999776 |

| Location | 19,555,902 – 19,555,996 |
|---|---|
| Length | 94 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 71.13 |
| Shannon entropy | 0.56299 |
| G+C content | 0.48844 |
| Mean single sequence MFE | -37.24 |
| Consensus MFE | -23.57 |
| Energy contribution | -21.38 |
| Covariance contribution | -2.18 |
| Combinations/Pair | 1.57 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.37 |
| SVM RNA-class probability | 0.999776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
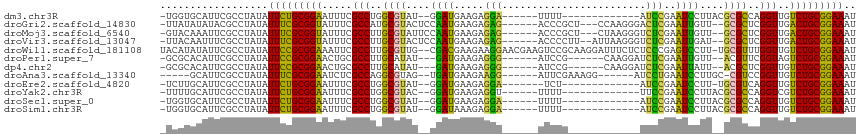
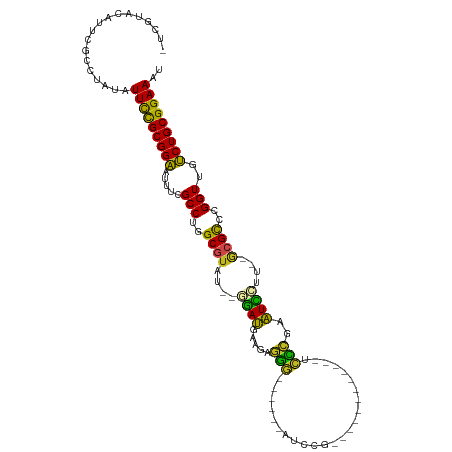
>dm3.chr3R 19555902 94 - 27905053 -UGGUGCAUUCGCCUAUAUUCUGCGGAAUUUCGCCUGGCGUAU--GGAUGAAGAGGA------UUUU-------------AUCCGAAUCCUUACGCGCCAGGUUGUCUGCGGAAAU -.((((....))))....(((((((((.....(((((((((..--((((.....(((------(...-------------))))..))))....)))))))))..))))))))).. ( -39.80, z-score = -4.14, R) >droGri2.scaffold_14830 4804845 104 - 6267026 -UUAUAUAUACGCCUAUAUUUCGCGGUAUUUCGCCAUGCGUACUCCAAUGAAGAGAG------ACCCGCU---CCAAGGGACUCGAAUUGUU--GCGCUCGGUUGACUGCGGAAAU -.................(((((((((.....(((..(((((...((((.....(((------.(((...---....))).)))..)))).)--))))..)))..))))))))).. ( -31.20, z-score = -1.54, R) >droMoj3.scaffold_6540 15493449 104 + 34148556 -GUACAAAUUCGCCUAUAUUUCGCGGUAUUUCGCCUUGCGUAUUCCAAUGAAGAGAG------ACCCGCU---CUAAGGGUCUCGAAUUGUU--GCGCUCGGUUGACUGCGGAAAU -.................(((((((((.....(((..(((((...((((.....(((------((((...---....)))))))..)))).)--))))..)))..))))))))).. ( -37.60, z-score = -3.30, R) >droVir3.scaffold_13047 4042400 105 + 19223366 -UUACAAUUUCGCCUAUAUUUCGCGGUAUUUCGCCUUGCGUACUCCAAUGAAGAGAG------ACCCCUU--AUUAAGGGUCUCGAAUUGAU--GCGCUCGGUUGACUGCGGAAAU -.................(((((((((.....(((..(((((...((((.....(((------((((...--.....)))))))..)))).)--))))..)))..))))))))).. ( -36.00, z-score = -3.43, R) >droWil1.scaffold_181108 1106657 113 + 4707319 UACAUAUAUUCGCCUAUAUUCCGCGGAAAUUCGCCUUGCGUUG--CGACGAAGAAGGAACGAAGUCCGCAAGGAUUUCUCUCCCGAGUCCUU-UGCGUUUGGUUGUCUGCGGAAAU ..................(((((((((.....(((..((((.(--.(((......(((..(((((((....)))))))..)))...))))..-.))))..)))..))))))))).. ( -41.50, z-score = -3.30, R) >droPer1.super_7 3157932 98 + 4445127 -GCGCACAUUCGCCUAUAUUCCGCGGAACUGCGCCUUGCAUAU---GAUGAAGAGGG------AUCCG------CAAGGAUCUCGAAUUGUU--ACGUUCGGUAGUCUGCGGAAAU -(((......))).....(((((((((.((((...........---........(((------((((.------...)))))))((((....--..)))).))))))))))))).. ( -37.40, z-score = -2.80, R) >dp4.chr2 3054999 98 - 30794189 -GCGCACAUUCGCCUAUAUUCCGCGGAACUGCGCCUUGCAUAU---GAUGAAGAGGG------AUCCG------CAAGGAUCUCGAAUUAUU--ACGCUCGGUUGUCUGCGGAAAU -(((......))).....(((((((((.....(((..((...(---(((((...(((------((((.------...)))))))...)))))--).))..)))..))))))))).. ( -38.40, z-score = -3.18, R) >droAna3.scaffold_13340 3721489 96 + 23697760 -----GCAUUCGCCUAUAUUUCGCGGAAUCUCGCCAGGCGUAG--UGAUGAAGAAGG------AUUCGAAAGG------AUCCUGAAUCCUUGC-CGUCCGGUUGUCUGCGGAAAU -----((....)).....(((((((((...(((...((((.((--.(((.....(((------((((....))------)))))..))))))))-)...)))...))))))))).. ( -35.90, z-score = -2.49, R) >droEre2.scaffold_4820 1981466 92 + 10470090 -UCUUGCAUUCGCCUAUAUUCUGCGGAAUUUCGCCUGGCGUAU--GGAUGAAGAGGA-------UCU-------------AUCCGAAUCCUU-UGCGUCAGGUUGUCUGCGGAAAU -....((....)).....(((((((((.....((((((((((.--((((.....(((-------...-------------.)))..))))..-))))))))))..))))))))).. ( -35.70, z-score = -3.52, R) >droYak2.chr3R 20461765 94 - 28832112 -UUUUGCAUUCGCCUAUAUUCUGCGGAAUUUCGCCUGGCGUAC--GGAUGAAGAGGU------UUUU-------------UUCCGAAUCCUUACGCGCCAGGUCGUCUGCGGAAAU -....((....)).....(((((((((.....(((((((((..--((((...((((.------...)-------------)))...))))....)))))))))..))))))))).. ( -34.90, z-score = -2.79, R) >droSec1.super_0 19914297 94 - 21120651 -UGGUGCAUUCGCCUAUAUUCUGCGGAAUUUCGCCUGGCGUAU--GGAUGAAGAGGA------UUUU-------------AUCCGAAUCCUUACGCGCCAGGUUGUCUGCGGAAAU -.((((....))))....(((((((((.....(((((((((..--((((.....(((------(...-------------))))..))))....)))))))))..))))))))).. ( -39.80, z-score = -4.14, R) >droSim1.chr3R 19385827 94 - 27517382 -UGGUGCAUUCGCCUAUAUUCUGCGGAAUUUCGCCUGGCGUAU--GGAUAAAGAGGA------UUUU-------------AUCCGAAUCCUUACGCGCCAGGUUGUCUGCGGAAAU -.((((....))))....(((((((((.....(((((((((..--((((.....(((------(...-------------))))..))))....)))))))))..))))))))).. ( -38.70, z-score = -4.04, R) >consensus _UCGUACAUUCGCCUAUAUUCCGCGGAAUUUCGCCUGGCGUAU__GGAUGAAGAGGG______AUCCG____________UCCCGAAUCCUU__GCGCCCGGUUGUCUGCGGAAAU ..................(((((((((.....(((..((((....((((.....(((........................)))..))))....))))..)))..))))))))).. (-23.57 = -21.38 + -2.18)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:38:47 2011