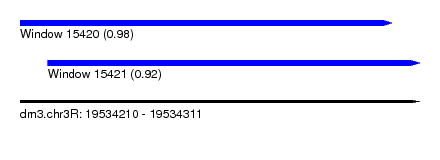
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,534,210 – 19,534,311 |
| Length | 101 |
| Max. P | 0.982188 |

| Location | 19,534,210 – 19,534,304 |
|---|---|
| Length | 94 |
| Sequences | 12 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 74.84 |
| Shannon entropy | 0.51386 |
| G+C content | 0.37429 |
| Mean single sequence MFE | -19.59 |
| Consensus MFE | -11.18 |
| Energy contribution | -10.73 |
| Covariance contribution | -0.45 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.982188 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
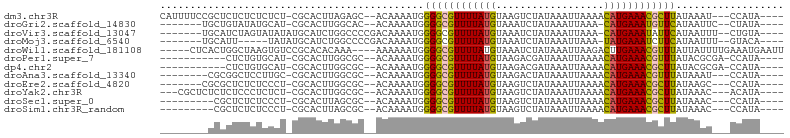
>dm3.chr3R 19534210 94 + 27905053 CAUUUUCCGCUCUCUCUCUCU-CGCACUUAGAGC--ACAAAAUGGGGCGUUUUAUGUAAGUCUAUAAAUUAAAACAUGAAACGCUUAUAAAU---CCAUA---- ((((((..(((((........-.......)))))--..))))))(((((((((((((................)))))))))))))......---.....---- ( -22.05, z-score = -4.12, R) >droGri2.scaffold_14830 4784424 87 + 6267026 -------UGCUGUAUAUGCAU-CGCACUUGGCAC--ACAAAAUGGGGCGUUUUAUGUAAAUCUAUAAAUUAAA-CAUGAAAUGUUCAUAAUUC--CUAUA---- -------(((((....(((..-.)))..))))).--........(((((((((((((...............)-)))))))))))).......--.....---- ( -15.46, z-score = -0.47, R) >droVir3.scaffold_13047 4022697 90 - 19223366 -------UGCAUCUAGUAUAUAUGCAUCUGGCCCCGACAAAAUGGGGCGUUUUAUGUAAAUCUAUAAAUUAAA-CAUGAAAUAUUCAUAAUUU--CUGUA---- -------(((((.........)))))....((((((......))))))(((((((((...............)-))))))))...........--.....---- ( -18.16, z-score = -1.36, R) >droMoj3.scaffold_6540 15470928 85 - 34148556 -------UGCAUU-----UAUAUGCAUCUGGCCCCGACAAAAUGGGGCGUUUUAUGUAAAUCUAUAAAUUAAA-UAUGAAAUCUUCAUAAUUU--GUACA---- -------(((((.-----...)))))....((((((......))))))..............(((((((....-(((((.....)))))))))--)))..---- ( -18.50, z-score = -1.82, R) >droWil1.scaffold_181108 1080691 95 - 4707319 -----CUCACUGGCUAAGUGUCCGCACACAAA----AAAAAAUGGGGCGUUUUAUGUAAAUCUAUAAAUUAAGACUUGAAACGUUUAUUAUUUUGAAAUGAAUU -----.(((........(((....))).((((----(.((....((((((((((.((.(((......)))...)).)))))))))).)).)))))...)))... ( -12.50, z-score = 0.38, R) >droPer1.super_7 3127945 85 - 4445127 -----------CUCUGUGCAU-CGCACUUGGCGC--ACAAAAUGGGGCGUUUUAUGUAAGACGAUAAAUUAAAACAUGAAACGUUUAUACGCGA-CCAUA---- -----------....((((..-.)))).((((((--.(.....)(((((((((((((................)))))))))))))....))).-)))..---- ( -21.39, z-score = -1.64, R) >dp4.chr2 3032121 85 + 30794189 -----------CUCUGUGCAU-CGCACUUGGCGC--ACAAAAUGGGGCGUUUUAUGUAAGACGAUAAAUUAAAACAUGAAACGCUUAUACGCGA-CCAUA---- -----------....((((..-.)))).((((((--.(.....)(((((((((((((................)))))))))))))....))).-)))..---- ( -23.89, z-score = -2.61, R) >droAna3.scaffold_13340 3700286 86 - 23697760 --------CGCGGCUCCUUGC-CGCACUUGGCGC--ACAAAAUGGGGCGUUUUAUGUAAGACUAUAAAUUAAAACAUGAAACGUUUAUAAAU---CCAUA---- --------.(((((.....))-)))..(((....--.))).((((((((((((((((................))))))))))).......)---)))).---- ( -24.10, z-score = -2.47, R) >droEre2.scaffold_4820 1959994 87 - 10470090 -------CGCGCUCUCUCCCU-CGCACUUGGCGC--ACAAAAUGGGGCGUUUUAUGUAAGUCUAUAAAUUAAAACAUGAAACGCUUAUAAGC---CCAUA---- -------.(((((........-.......)))))--.....((((((((((((((((................))))))))))).......)---)))).---- ( -23.06, z-score = -2.30, R) >droYak2.chr3R 20432023 91 + 28832112 ---CGCUCUCUCUCCCUCUCU-CGCACUUGGCGC--ACAAAAUGGGGCGUUUUAUGUAAGUCUAUAAAUUAAAACAUGAAACGCUUAUAAAC---ACAUA---- ---..........(((.....-(((.....))).--.......)))(((((((((((................)))))))))))........---.....---- ( -18.21, z-score = -2.44, R) >droSec1.super_0 19892676 85 + 21120651 ---------CGCUCUCUCCCU-CGCACUUAGCGC--ACAAAAUGGGGCGUUUUAUGUAAGUCUAUAAAUUAAAACAUGAAACGCUUAUAAAC---CCAUA---- ---------............-(((.....))).--.....((((((((((((((((................))))))))))).......)---)))).---- ( -18.90, z-score = -2.54, R) >droSim1.chr3R_random 961027 85 + 1307089 ---------CGCUCUCUCCCU-CGCACUUAGCGC--ACAAAAUGGGGCGUUUUAUGUAAGUCUAUAAAUUAAAACAUGAAACGCUUAUAAAC---CCAUA---- ---------............-(((.....))).--.....((((((((((((((((................))))))))))).......)---)))).---- ( -18.90, z-score = -2.54, R) >consensus ________GCUCUCUCUCCAU_CGCACUUGGCGC__ACAAAAUGGGGCGUUUUAUGUAAGUCUAUAAAUUAAAACAUGAAACGCUUAUAAAC___CCAUA____ ............................................((((((((((((..................)))))))))))).................. (-11.18 = -10.73 + -0.45)
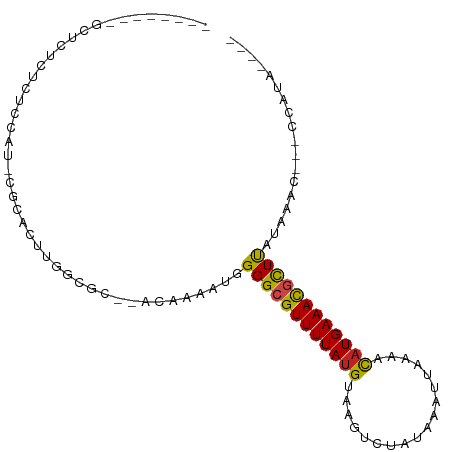
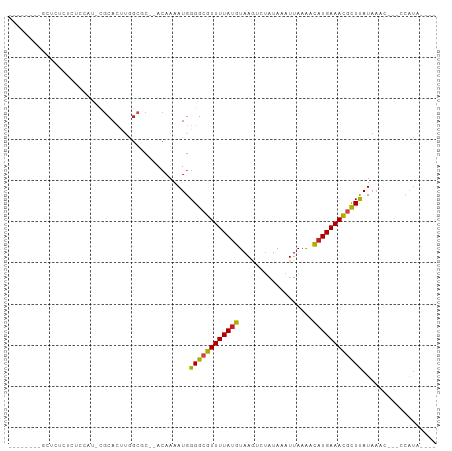
| Location | 19,534,217 – 19,534,311 |
|---|---|
| Length | 94 |
| Sequences | 12 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 75.34 |
| Shannon entropy | 0.52453 |
| G+C content | 0.38081 |
| Mean single sequence MFE | -20.16 |
| Consensus MFE | -11.18 |
| Energy contribution | -10.73 |
| Covariance contribution | -0.45 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.919069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 19534217 94 + 27905053 ----CGCUCUCUCUCUCU-CGCACUUAGAGC--ACAAAAUGGGGCGUUUUAUGUAAGUCUAUAAAUUAAAACAUGAAACGCUUAUAAAU--CCAUAUGACUGC-- ----.(((((........-.......)))))--.....((((((((((((((((................)))))))))))).......--))))........-- ( -20.06, z-score = -2.46, R) >droGri2.scaffold_14830 4784424 94 + 6267026 ----UGCUGUAUAUGCAU-CGCACUUGGCAC--ACAAAAUGGGGCGUUUUAUGUAAAUCUAUAAAUUAAA-CAUGAAAUGUUCAUAAUUC-CUAUAUGACCAA-- ----(((((....(((..-.)))..))))).--......(((.(((((((((((...............)-))))))))))(((((....-...)))))))).-- ( -16.46, z-score = -0.15, R) >droVir3.scaffold_13047 4022697 97 - 19223366 ----UGCAUCUAGUAUAUAUGCAUCUGGCCCCGACAAAAUGGGGCGUUUUAUGUAAAUCUAUAAAUUAAA-CAUGAAAUAUUCAUAAUUU-CUGUAUGACCAA-- ----..(((.(((....((((......((((((......))))))(((((((((...............)-))))))))...))))....-))).))).....-- ( -19.76, z-score = -1.27, R) >droMoj3.scaffold_6540 15470928 92 - 34148556 ----UGCAUUU-----AUAUGCAUCUGGCCCCGACAAAAUGGGGCGUUUUAUGUAAAUCUAUAAAUUAAA-UAUGAAAUCUUCAUAAUUU-GUACAUGACCAA-- ----(((((..-----..)))))....((((((......))))))...(((((((...........((((-(((((.....)))).))))-))))))))....-- ( -19.70, z-score = -1.63, R) >droWil1.scaffold_181108 1080691 98 - 4707319 ------CUCACUGGCUAAGUGUCCGCACACAAAAAAAAAUGGGGCGUUUUAUGUAAAUCUAUAAAUUAAGACUUGAAACGUUUAUUAUUU-UGAAAUGAAUUUGA ------.(((........(((....))).(((((.((....((((((((((.((.(((......)))...)).)))))))))).)).)))-))...)))...... ( -12.50, z-score = 0.74, R) >droPer1.super_7 3127945 92 - 4445127 --------CUCUGUGCAU-CGCACUUGGCGC--ACAAAAUGGGGCGUUUUAUGUAAGACGAUAAAUUAAAACAUGAAACGUUUAUACGCGACCAUAUGACGAC-- --------.........(-((((..((((((--.(.....)(((((((((((((................)))))))))))))....))).)))..)).))).-- ( -23.29, z-score = -1.90, R) >dp4.chr2 3032121 92 + 30794189 --------CUCUGUGCAU-CGCACUUGGCGC--ACAAAAUGGGGCGUUUUAUGUAAGACGAUAAAUUAAAACAUGAAACGCUUAUACGCGACCAUAUGACGAC-- --------.........(-((((..((((((--.(.....)(((((((((((((................)))))))))))))....))).)))..)).))).-- ( -25.79, z-score = -2.93, R) >droAna3.scaffold_13340 3700286 93 - 23697760 -----CGCGGCUCCUUGC-CGCACUUGGCGC--ACAAAAUGGGGCGUUUUAUGUAAGACUAUAAAUUAAAACAUGAAACGUUUAUAAAU--CCAUAUGACUAG-- -----.(((((.....))-)))..(((....--.))).((((((((((((((((................))))))))))).......)--))))........-- ( -24.10, z-score = -2.24, R) >droEre2.scaffold_4820 1959994 94 - 10470090 ----CGCGCUCUCUCCCU-CGCACUUGGCGC--ACAAAAUGGGGCGUUUUAUGUAAGUCUAUAAAUUAAAACAUGAAACGCUUAUAAGC--CCAUAUGACUGU-- ----.(((((........-.......)))))--.....((((((((((((((((................))))))))))).......)--))))........-- ( -23.06, z-score = -1.62, R) >droYak2.chr3R 20432023 98 + 28832112 CGCUCUCUCUCCCUCUCU-CGCACUUGGCGC--ACAAAAUGGGGCGUUUUAUGUAAGUCUAUAAAUUAAAACAUGAAACGCUUAUAAAC--ACAUAUGACUGU-- .((..((...(((.....-(((.....))).--.......)))(((((((((((................)))))))))))........--......))..))-- ( -18.61, z-score = -1.55, R) >droSec1.super_0 19892676 92 + 21120651 ------CGCUCUCUCCCU-CGCACUUAGCGC--ACAAAAUGGGGCGUUUUAUGUAAGUCUAUAAAUUAAAACAUGAAACGCUUAUAAAC--CCAUAUGACUGU-- ------.((..((.....-(((.....))).--.....((((((((((((((((................))))))))))).......)--))))..))..))-- ( -19.30, z-score = -1.87, R) >droSim1.chr3R_random 961027 92 + 1307089 ------CGCUCUCUCCCU-CGCACUUAGCGC--ACAAAAUGGGGCGUUUUAUGUAAGUCUAUAAAUUAAAACAUGAAACGCUUAUAAAC--CCAUAUGACUGU-- ------.((..((.....-(((.....))).--.....((((((((((((((((................))))))))))).......)--))))..))..))-- ( -19.30, z-score = -1.87, R) >consensus ______CUCUCUCUCCAU_CGCACUUGGCGC__ACAAAAUGGGGCGUUUUAUGUAAGUCUAUAAAUUAAAACAUGAAACGCUUAUAAAC__CCAUAUGACUAU__ .........................................((((((((((((..................))))))))))))...................... (-11.18 = -10.73 + -0.45)
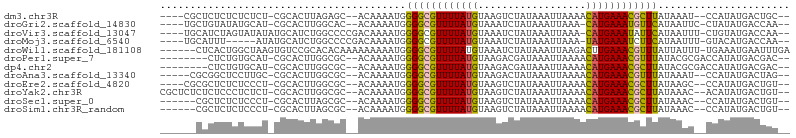

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:38:44 2011