
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,496,847 – 19,496,937 |
| Length | 90 |
| Max. P | 0.597742 |

| Location | 19,496,847 – 19,496,937 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 75.56 |
| Shannon entropy | 0.46326 |
| G+C content | 0.35174 |
| Mean single sequence MFE | -18.91 |
| Consensus MFE | -8.68 |
| Energy contribution | -8.26 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.597742 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
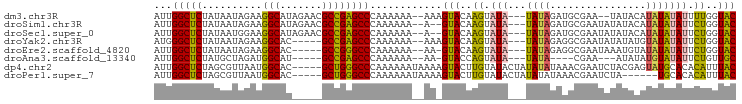
>dm3.chr3R 19496847 90 + 27905053 AUUGGCUCUAUAAUAGAAGGCAUAGAACGCCGAGCCCAAAAAA--AAAGUACAAGUAUA---UAUAGAUGCGAA--UAUACAUAUAUAUUUUGGUAC ...(((((..........(((.......)))))))).......--...((((.((((((---(((..(((....--)))..)))))))))...)))) ( -17.10, z-score = -1.77, R) >droSim1.chr3R 19326518 90 + 27517382 AUUGGCUCUAUAAUAGAAGGCAUAGAACGCCGAGCCCAAAAAA--A--GUACAAGUAUA---UAUAGAUGCGAAUAUAUACAUAUAUAUUCUGGUAC ...(((((..........(((.......)))))))).......--.--((((..((((.---.....))))((((((((....))))))))..)))) ( -19.70, z-score = -2.22, R) >droSec1.super_0 19853912 90 + 21120651 AUUGGCUCUAUAAUGGAAGGCAUAGAACGCCGAGCCCAAAAAA--A--GUACAAGUAUA---UAUAGAUGCGAAUAUAUACAUAUAUAUUCUGGUAC ...(((((..........(((.......)))))))).......--.--((((..((((.---.....))))((((((((....))))))))..)))) ( -19.70, z-score = -1.83, R) >droYak2.chr3R 20393494 87 + 28832112 AUGGGCUCUAUAAUAGAAGGCAC-----GCCGAGCCCAAAAAA--AAAGUACAAGUAUA---UAUAGAGGCGAAUAUAUAUGUAUAUAUUCUGGUAC .(((((((..........((...-----.))))))))).....--...((((.((((((---((((..............))))))))))...)))) ( -20.74, z-score = -2.13, R) >droEre2.scaffold_4820 1922067 86 - 10470090 AUUGGCUCUAUAAUAGAAGGCAC-----GCCGGGCCCAAAAAA--AA-GUACAAGUAUA---UAUAGAGGCGAAUAAAUGUAUAUAUAUUCUGGUAC ...(((((..........((...-----.))))))).......--..-((((.((.(((---((((...(((......))).))))))).)).)))) ( -15.70, z-score = -0.32, R) >droAna3.scaffold_13340 3662639 79 - 23697760 AUUGGCUCUAUGCUAGAUGGCAU-----GCCGAGCCCAAAAAA--AA-GUACCAGUAUA---UAUA----CGAA---AUAUAUGUAUAUUCUGUUGC ...(((((((((((....)))))-----)..))))).......--..-(((.(((...(---((((----((..---.....))))))).))).))) ( -20.40, z-score = -2.42, R) >dp4.chr2 2996576 92 + 30794189 AUUGGCUCUAGCGUUAAUGGCAC-----GCUGGGCCCAAAAAAUAAAAGUACUUGUAUACUAUAUAUAAACGAAUCUACGAGUAUGCACACAUUUAC .((((..(((((((.......))-----)))))..)))).........(((((((((...................)))))))))............ ( -21.61, z-score = -2.17, R) >droPer1.super_7 3092139 86 - 4445127 AUUGGCUCUAGCGUUAAUGGCAC-----GCUGGGCCCAAAAAAUAAAAGUACUUGUAUACUAUAUAUAAACGAAUCUA------UGCACACAUUUAC .((((..(((((((.......))-----)))))..))))....((((.((...((((((.................))------)))).)).)))). ( -16.33, z-score = -0.99, R) >consensus AUUGGCUCUAUAAUAGAAGGCAC_____GCCGAGCCCAAAAAA__AA_GUACAAGUAUA___UAUAGAUGCGAAUAUAUACAUAUAUAUUCUGGUAC ...(((((..........(((.......))))))))............................................................. ( -8.68 = -8.26 + -0.42)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:38:42 2011