| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,434,301 – 8,434,397 |
| Length | 96 |
| Max. P | 0.534984 |

| Location | 8,434,301 – 8,434,397 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 65.06 |
| Shannon entropy | 0.73384 |
| G+C content | 0.58709 |
| Mean single sequence MFE | -35.46 |
| Consensus MFE | -12.80 |
| Energy contribution | -12.72 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.534984 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
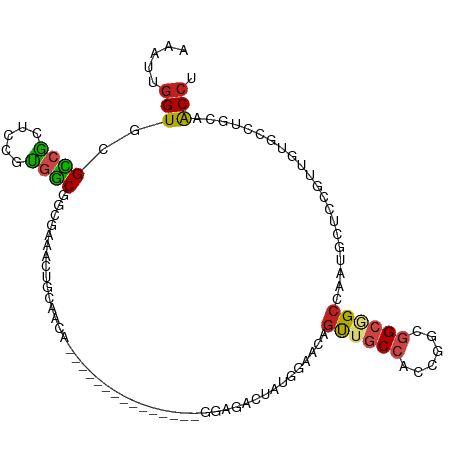
>dm3.chr2L 8434301 96 - 23011544 AAAUUGGUGCGCCGCUCCUUGGCGGCGAAACUGCAACA---------------GGAGACUUUGGAACAGUUGCCACCGGCGGCGGCCAAUGCUCCGUUGUGUCUCCAACCU ...((((.((((((((....))))))).....(((((.---------------((((...((((....((((((...))))))..))))..)))))))))..).))))... ( -39.70, z-score = -1.93, R) >droSim1.chr2L 8220376 96 - 22036055 AAAUUGGUGCGCCGCUCCUUGGCGGCGAAACUGCAACA---------------GGAGACUUUGGAACAGUUGCCACCGGCGGCGGCCAAUGCUCCGUUGUGUCUCCAACCU ...((((.((((((((....))))))).....(((((.---------------((((...((((....((((((...))))))..))))..)))))))))..).))))... ( -39.70, z-score = -1.93, R) >droSec1.super_3 3930571 96 - 7220098 AAAUUGGUGCGCCGCUCCUUGGCGGCGAAACUGCAACA---------------GGAGACUUUGGAACAGUUGCCACCGGCGGCGGCCAAUGCUCCGUUGUGUCUCCAACCU ...((((.((((((((....))))))).....(((((.---------------((((...((((....((((((...))))))..))))..)))))))))..).))))... ( -39.70, z-score = -1.93, R) >droYak2.chr2L 11072222 96 - 22324452 AGAUUGGUACGCCGCUCAUUGGCGGCGAAACUGCAACA---------------GGAGACUUUGGAACAGUUACCACCGGCGGCGGCCAAUGCUUCAUUGUGUCUCCUACCU .....(((.(((((((....)))))))..........(---------------((((((..((((.((....((.((...)).))....)).))))....)))))))))). ( -33.50, z-score = -1.55, R) >droEre2.scaffold_4929 17342913 96 - 26641161 AGAUUGGUUCGCCGCUCCUUGGCGGCGAAGCUGCAACA---------------GGAGACUUUGGAACAGUUGCCACCGGCGGCGGCCAAUGCUCCGUUGUUUCUCCAACCU ((.(((((((((((((....)))))))))(..(((((.---------------((((...((((....((((((...))))))..))))..)))))))))..).)))).)) ( -42.20, z-score = -2.64, R) >droAna3.scaffold_12916 10089247 96 - 16180835 AAAUUGGUUCGCCGCUCGGUGGCGGCUAAGCUCCAGCA---------------CGAUUCUGUGGAGGAGCUGCCCCCGGCGGCGGCGAAUGCUUCCCUGUGCCUGCUACCU ......(((((((((.((.(((.(((..((((((..((---------------((....))))..))))))))).))).)))))))))))((........))......... ( -44.20, z-score = -1.82, R) >dp4.chr4_group1 1201191 96 + 5278887 CGACUGGUGCGCCGUUCUGUGGCAGUAAAGCUCCACCA---------------GGACACCAUUGAACAGCUGCCGCCGCCCGCCGCCAAUCUGCCGCUCUGCCUGCAGCCA .((.(((((.((.(..(.((((((((.....(((....---------------)))((....))....)))))))).)..)))))))).)).((.((.......)).)).. ( -29.10, z-score = 0.84, R) >droPer1.super_5 1187057 96 - 6813705 CGACUGGUGCGCCGUUCUGUGGCAGUAAAGCUCCACCA---------------GGACACCAUUGAACAGCUGCCGCCGCCCGCCGCCAAUGUGCCGCUCUGCCUGCAGCCA ....(((((.((.(..(.((((((((.....(((....---------------)))((....))....)))))))).)..)))))))).((.((.((.......)).)))) ( -27.90, z-score = 1.59, R) >droWil1.scaffold_181132 325845 108 - 1035393 AAACUAGUGCGUCAAUCGGUGGCCACCAAGUUGCAGCAGCAGCAUCAACAUAUGGAUACAAUC---CUGUUACCUCAGGCGGCUGCAAAUUCCGAUAUGUUAUUGCAGCCG ...((((((.((((.....)))))))...(((((....))))).........))).......(---(((......))))(((((((((..............))))))))) ( -30.14, z-score = -0.59, R) >droVir3.scaffold_12723 3394317 108 + 5802038 AAGUUGGUGAGACGCGCCGUCUCAACCAAAUUGCGUCAACAGCAACAA---CCGCAUGCAAUAGAGACGUUGCCUACUGCGGCAGCCAAUAUAGAACUAAACCUACUACCC ..((((((((((((...)))))).......((((.......))))...---(((((.(((((......)))))....)))))..))))))..................... ( -27.50, z-score = -1.62, R) >droMoj3.scaffold_6500 16781707 108 + 32352404 AAGUUGGUCCGUCGCUCCGCUGCAACCAAACUGCUGCAGCAGCAACAG---UCGCAUGCAAUCGAGACGCUGCCACCGCCGGCGGCUAAUCUGGAAAUGACCUUGCUACCU .(((.((((.((.(((..((((((.(......).))))))))).))..---..........((.(((.((((((......))))))...))).))...))))..))).... ( -34.70, z-score = -0.48, R) >droGri2.scaffold_15252 1911914 111 - 17193109 AAGUUGGUGCGUCGCGCCGUGGCUGCCAAACUGCAGCAACAACAGCAAUCGCCGCAUACGAUAGAUGCACUGCCUCCGGCGGCAGCCAAUGUAGAAGUGACCUUGCUACCC ..(((((((((((..((.((.(((((......)))))....)).)).((((.......)))).))))))(((((......))))))))))(((((((....))).)))).. ( -37.20, z-score = -0.13, R) >consensus AAAUUGGUGCGCCGCUCCGUGGCGGCGAAACUGCAACA_______________GGAGACUAUGGAACAGUUGCCACCGGCGGCGGCCAAUGCUCCGUUGUGCCUGCAACCU .....(((..((((.....)))).............................................((((((......)))))).....................))). (-12.80 = -12.72 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:26:02 2011