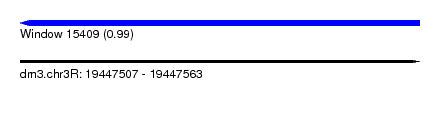
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,447,507 – 19,447,563 |
| Length | 56 |
| Max. P | 0.990559 |

| Location | 19,447,507 – 19,447,563 |
|---|---|
| Length | 56 |
| Sequences | 5 |
| Columns | 57 |
| Reading direction | reverse |
| Mean pairwise identity | 58.41 |
| Shannon entropy | 0.70697 |
| G+C content | 0.48620 |
| Mean single sequence MFE | -13.72 |
| Consensus MFE | -8.20 |
| Energy contribution | -7.20 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 2.00 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.990559 |
| Prediction | RNA |
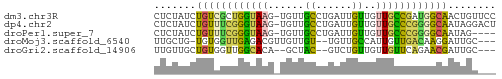
Download alignment: ClustalW | MAF
>dm3.chr3R 19447507 56 - 27905053 CUCUAUCUGUCGCUGGUAAG-UGUUGCCUGAUUGUUGUUGCCGAUGGCAACUGUUCC ........((((..((((..-...))))))))....((((((...))))))...... ( -10.60, z-score = -0.52, R) >dp4.chr2 2940038 56 - 30794189 CUCUAUCUGUUUCGGGUAAG-UGUUGCCUGAUUGUUGUUGCCCGGGGCAAUAGGACU .(((((.(((..(((((((.-....((......))..)))))))..))))))))... ( -18.50, z-score = -2.15, R) >droPer1.super_7 3035589 52 + 4445127 CUCUAUCUGUUUCGGGUAAG-UGUUGCCUGAUUGUUGUUGCCCGGGGCAAUAG---- ..((((.(((..(((((((.-....((......))..)))))))..)))))))---- ( -16.70, z-score = -2.55, R) >droMoj3.scaffold_6540 2545209 51 + 34148556 UUGCUG-UGUGGUUGAGACGUUGUUGU--UGUUGCCAUUGUUGACAAGGAUUGC--- ((((..-.(((((...((((....)))--)...)))))....).))).......--- ( -9.00, z-score = 0.12, R) >droGri2.scaffold_14906 9972669 50 + 14172833 UUGUUGCUGUGGUUGGCACA--GCUAC--GUCUGUUGUUGUUCAGAACGAUUGC--- .....((((((.....))))--))..(--(((((........))).))).....--- ( -13.80, z-score = -0.80, R) >consensus CUCUAUCUGUGGCUGGUAAG_UGUUGCCUGAUUGUUGUUGCCCAGGGCAAUUGC___ .......((((((((((((......((......))..))))))))))))........ ( -8.20 = -7.20 + -1.00)
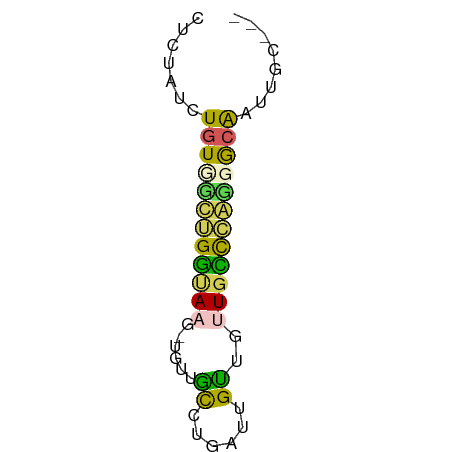
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:38:34 2011