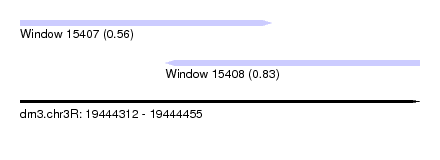
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,444,312 – 19,444,455 |
| Length | 143 |
| Max. P | 0.830606 |

| Location | 19,444,312 – 19,444,402 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 72.89 |
| Shannon entropy | 0.52810 |
| G+C content | 0.53135 |
| Mean single sequence MFE | -15.26 |
| Consensus MFE | -7.69 |
| Energy contribution | -8.16 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.555538 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
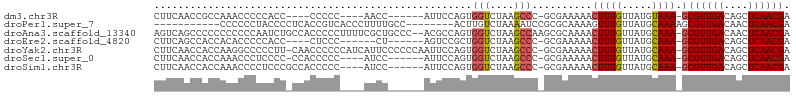
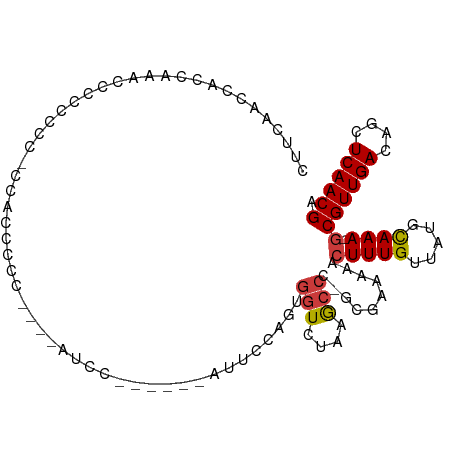
>dm3.chr3R 19444312 90 + 27905053 CUUCAACCGCCAAACCCCCACC----CCCCC----AACC------AUUCCAGUGGUCUAAGCCC-GCGAAAAACUUUGUUAUGUAAA-GCGUUGACAGCUCAACGA .......(((.......((((.----.....----....------......)))).........-))).....(((((.....))))-)((((((....)))))). ( -11.28, z-score = -0.07, R) >droPer1.super_7 3032089 87 - 4445127 -----------CCCCCCUACCCCUCACCGUCACCCUUUUGCC--------ACUUGUCUAAAAUCCGCGCAAAAGUUUGUUAUGCAAAAGCGUUGACAACUCAACGA -----------................(((....((((((((--------...............).))))))).(((((((((....))).))))))....))). ( -12.56, z-score = -1.66, R) >droAna3.scaffold_13340 3608050 103 - 23697760 AGUCAGCCCCCCCCCCCAAUCUGCCACCCCCUUUUCGCUGCCC--ACGCCAGUGGUCUAAGCCAAGCGCAAAACUUUGUUAUGCAAA-GCGUUGACAACUCAACGA .((((((........................((((((((((((--((....)))).....))..)))).))))(((((.....))))-).)))))).......... ( -21.30, z-score = -1.60, R) >droEre2.scaffold_4820 1868173 88 - 10470090 CUUCAGCCACCACACCCCCACC----CUCCC------CU------AGUCCGCUGGUCUAAGCCC-GCGAAAAACUUUGUUAUGCAAA-GCGUUGACAGCUCAACGA ......................----.....------..------....(((.(((....))).-))).....(((((.....))))-)((((((....)))))). ( -16.60, z-score = -1.19, R) >droYak2.chr3R 20338047 103 + 28832112 CUUCAACCACCAAGGCCCCCUU-CAACCCCCCAUCAUUCCCCCCAAUUCCAGUGGUCUAAGCCC-GCGAAAAACUUUGUUAUGCAAA-GCGUUGACAGCUCAACGA .(((.......((((...))))-............................((((.......))-)))))...(((((.....))))-)((((((....)))))). ( -15.20, z-score = -0.58, R) >droSec1.super_0 19801815 93 + 21120651 CUUCAACCACCAAACCCUCCCC-CCACCCCC----AUCC------AUUCCAGUGGUCUAAGCCC-GCGAAAAACUUUGUUAUGCAAA-GCGUUGACAGCUCAACGA (((..(((((............-........----....------......)))))..)))...-........(((((.....))))-)((((((....)))))). ( -13.91, z-score = -1.49, R) >droSim1.chr3R 19273762 94 + 27517382 CUUCAACCACCAAACCCCUCCCGCCACCCCC----AUCC------AUUCCAGUGGUCUAAGCCC-GCGAAAAACUUUGUUAUGCAAA-GCGUUGACAGCUCAACGA .....................(((.......----..((------((....)))).........-))).....(((((.....))))-)((((((....)))))). ( -15.97, z-score = -1.70, R) >consensus CUUCAACCACCAAACCCCCCCC_CCACCCCC____AUCC______AUUCCAGUGGUCUAAGCCC_GCGAAAAACUUUGUUAUGCAAA_GCGUUGACAGCUCAACGA ............................................................((...(((((....)))))...)).....((((((....)))))). ( -7.69 = -8.16 + 0.47)

| Location | 19,444,364 – 19,444,455 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 91.21 |
| Shannon entropy | 0.15867 |
| G+C content | 0.45640 |
| Mean single sequence MFE | -28.38 |
| Consensus MFE | -21.86 |
| Energy contribution | -22.70 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.830606 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
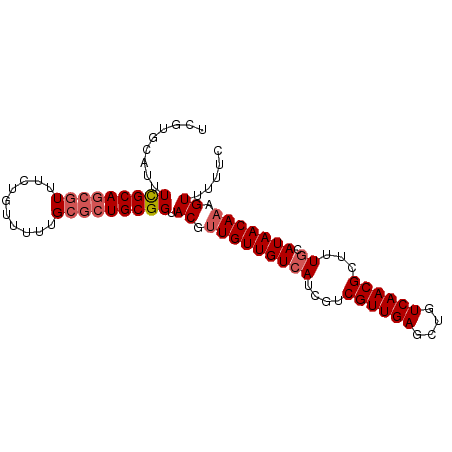
>dm3.chr3R 19444364 91 - 27905053 UCGUGCAUUUCGCAGCGUUUCUGUUUUUGCGCUGCGGUACGUUGUUGUCAUCGUCGUUGAGCUGUCAACGCUUUACAUAACAAAGUUUUUC .(((((....((((((((..........)))))))))))))(((((((......((((((....))))))......)))))))........ ( -28.00, z-score = -2.36, R) >droAna3.scaffold_13340 3608115 83 + 23697760 ------UUUUUGCAACGUUGCUGCUCUUG--CUGCGGUACGCUGUUGUCAACGUCGUUGAGUUGUCAACGCUUUGCAUAACAAAGUUUUGC ------.....(((((((.(((((.....--..))))))))..((((.((((........)))).))))((((((.....)))))).)))) ( -23.10, z-score = -0.34, R) >droEre2.scaffold_4820 1868223 91 + 10470090 UCGUGCAUUUCGCAGCGUUUCUGUUUUUGCGCUGCGGUACGUUGUUGUCAUCGUCGUUGAGCUGUCAACGCUUUGCAUAACAAAGUUUUUC .(((((....((((((((..........)))))))))))))(((((((((..(.((((((....)))))))..)).)))))))........ ( -30.70, z-score = -2.92, R) >droSec1.super_0 19801870 91 - 21120651 UCGUGCAUUUCGCAGCGUUUCUGUUUUUGCGCUGCGGUACGUUGUUGUCAUCAUCGUUGAGCUGUCAACGCUUUGCAUAACAAAGUUUUUC .(((((....((((((((..........)))))))))))))(((((((((....((((((....))))))...)).)))))))........ ( -29.40, z-score = -2.69, R) >droSim1.chr3R 19273818 91 - 27517382 UCGUGCAUUUCGCAGCGUUUCUGUUUUUGCGCUGCGGUACGUUGUUGUCAUCGUCGUUGAGCUGUCAACGCUUUGCAUAACAAAGUUUUUC .(((((....((((((((..........)))))))))))))(((((((((..(.((((((....)))))))..)).)))))))........ ( -30.70, z-score = -2.92, R) >consensus UCGUGCAUUUCGCAGCGUUUCUGUUUUUGCGCUGCGGUACGUUGUUGUCAUCGUCGUUGAGCUGUCAACGCUUUGCAUAACAAAGUUUUUC .........(((((((((..........))))))))).((.(((((((((....((((((....))))))...)).))))))).))..... (-21.86 = -22.70 + 0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:38:33 2011