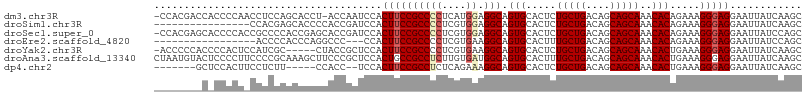
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,403,412 – 19,403,557 |
| Length | 145 |
| Max. P | 0.763511 |

| Location | 19,403,412 – 19,403,522 |
|---|---|
| Length | 110 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 68.76 |
| Shannon entropy | 0.62499 |
| G+C content | 0.58342 |
| Mean single sequence MFE | -23.50 |
| Consensus MFE | -9.92 |
| Energy contribution | -10.51 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.714856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
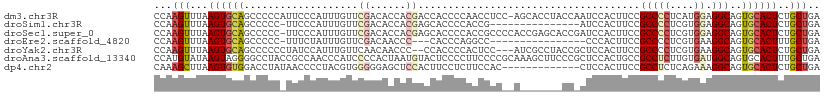
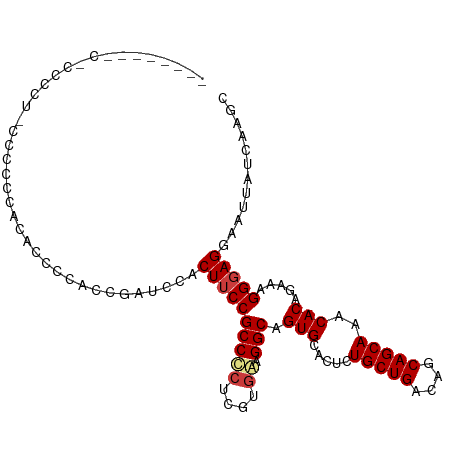
>dm3.chr3R 19403412 110 + 27905053 CCAAGUUUAAGUGCAGCCCCCAUUCCCAUUUGUUCGACACCACGACCACCCCAACCUCC-AGCACCUACCAAUCCACUUCCGCCCCUCAUGGAGGCAGUGCACUCUGCUGA ...(((...((((((((.............((.(((......))).))...........-.))..................(((((....)).)))..))))))..))).. ( -19.91, z-score = -1.02, R) >droSim1.chr3R 19225463 95 + 27517382 CCAAGUUUAAGUGCAGCCCCC-UUCCCAUUUGUUCGACACCACGAGCACCCCACCG---------------AUCCACUUCCGCCCCUCGUGGAGGCAGUGCACUCUGCUGA ...(((...(((((((((...-........((((((......))))))........---------------.......(((((.....))))))))..))))))..))).. ( -25.10, z-score = -2.03, R) >droSec1.super_0 19762494 110 + 21120651 CCAAGUUUAAGUGCAGCCCCC-UUCCCAUUUGUUCGACACCACGAGCACCCCACCGCCCCACCGAGCACCGAUCCACUUCCGCCCCUCGUGGAGGCAGUGCACUCUGCUGA ...(((...((((((......-........((((((......)))))).......(((((((.(((...((.........))...))))))).)))..))))))..))).. ( -30.20, z-score = -2.71, R) >droEre2.scaffold_4820 1829433 91 - 10470090 CCAAGUUUAAGUGCAGCCCCC-UUUCUAUUUGUUCGACAACCC---CACCCAGGCC----------------CCCACUUCCGCCCCUCGUGAAGGCAGUGCACUUUGCUGA ...(((..((((((((((...-.......(((.....)))...---(((...(((.----------------.........)))....)))..)))..))))))).))).. ( -18.60, z-score = -0.73, R) >droYak2.chr3R 20289305 106 + 28832112 CCAAGUUUAAGUGCAGCCCCCCUAUCCAUUUGUUCAACAACCC--CCACCCCACUCC---AUCGCCUACCGCUCCACUUCCGCCCCUCGUGAAGGCAGUGCACUCUGCUGA ...(((...(((((...............(((.....)))...--.......(((((---.((((.....((.........)).....)))).)).))))))))..))).. ( -16.30, z-score = -0.76, R) >droAna3.scaffold_13340 3553328 111 - 23697760 CCAUGUAUAAGUAGGGGCCUACCGCCAACCCAUCCCCACUAAUGUACUCCCCUUCCCCGCAAAGCUUCCCGCUCCACUGCCGCCUCUUGUGAUGGCAGUGCACUUUGCUGA ....((((((((.((((................)))))))..)))))...........(((((((.....))..((((((((.(......).))))))))...)))))... ( -27.29, z-score = -1.18, R) >dp4.chr2 2890654 98 + 30794189 CAAAGCUUAAGUGUGGACCUAUAACCCCUACGUGGGGGAGCUCCACUUCCUCUUCCAC-------------CUCCACUUCCGCCUCUCAGAAAGGCAGUGCACUCUGCUGA ...(((...((((((((.((....((((.....)))).)).)))))............-------------...((((...((((.......)))))))).)))..))).. ( -27.10, z-score = -0.57, R) >consensus CCAAGUUUAAGUGCAGCCCCC_UUCCCAUUUGUUCGACACCACGACCACCCCACCCCC__A_______CC_AUCCACUUCCGCCCCUCGUGAAGGCAGUGCACUCUGCUGA ...(((...((((((...................((......)).....................................(((((....)).)))..))))))..))).. ( -9.92 = -10.51 + 0.60)
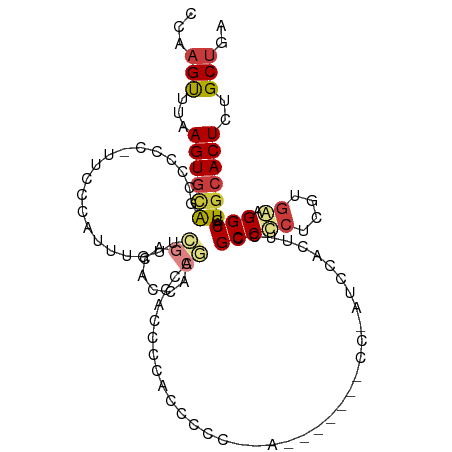
| Location | 19,403,451 – 19,403,557 |
|---|---|
| Length | 106 |
| Sequences | 7 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 75.62 |
| Shannon entropy | 0.48621 |
| G+C content | 0.57532 |
| Mean single sequence MFE | -26.67 |
| Consensus MFE | -18.34 |
| Energy contribution | -18.67 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.06 |
| Mean z-score | -0.95 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.763511 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 19403451 106 + 27905053 -CCACGACCACCCCAACCUCCAGCACCU-ACCAAUCCACUUCCGCCCCUCAUGGAGGCAGUGCACUCUGCUGACAGCAGCAAACACAGAAAGGGAGGAAUUAUCAAGC -...............(((((.......-..............(((((....)).))).(((....((((.....))))....)))......)))))........... ( -25.30, z-score = -1.01, R) >droSim1.chr3R 19225501 92 + 27517382 ----------------CCACGAGCACCCCACCGAUCCACUUCCGCCCCUCGUGGAGGCAGUGCACUCUGCUGACAGCAGCAAACACAGAAAGGGAGGAAUUAUCAAGC ----------------................(((...((((((((((....)).))).(((....((((.....))))....))).....))))).....))).... ( -23.60, z-score = -0.12, R) >droSec1.super_0 19762532 107 + 21120651 -CCACGAGCACCCCACCGCCCCACCGAGCACCGAUCCACUUCCGCCCCUCGUGGAGGCAGUGCACUCUGCUGACAGCAGCAAACACAGAAAGGGAGGAAUUAUCCAGC -......((.((((((.(((((((.(((...((.........))...))))))).))).))).....(((((....)))))..........))).(((....))).)) ( -30.60, z-score = -1.15, R) >droEre2.scaffold_4820 1829471 88 - 10470090 -----------------ACCCCACCCAGGCCC---CCACUUCCGCCCCUCGUGAAGGCAGUGCACUUUGCUGACAGCAGCAAACACAGAAAGGGAGGAAUUAUCCAGC -----------------...((.(((..(((.---.(((...........)))..))).(((...(((((((....)))))))))).....))).))........... ( -24.40, z-score = -0.79, R) >droYak2.chr3R 20289344 102 + 28832112 -ACCCCCACCCCACUCCAUCGC-----CUACCGCUCCACUUCCGCCCCUCGUGAAGGCAGUGCACUCUGCUGACAGCAGCAAACACUGAAAGGGAGGAAUUAUCAAGC -.(((((........((.((((-----.....((.........)).....)))).))(((((....((((.....))))....)))))...))).))........... ( -24.40, z-score = -0.54, R) >droAna3.scaffold_13340 3553366 108 - 23697760 CUAAUGUACUCCCCUUCCCCGCAAAGCUUCCCGCUCCACUGCCGCCUCUUGUGAUGGCAGUGCACUUUGCUGACAGCAGCAAACACUGAAAGGGAGGAAUUAUCAAGC .((((...(((((.(((...((..(((.....)))..(((((((.(......).)))))))))..(((((((....)))))))....))).)))))..))))...... ( -33.70, z-score = -2.03, R) >dp4.chr2 2890693 94 + 30794189 -------GCUCCACUUCCUCUU-----CCACC--UCCACUUCCGCCUCUCAGAAAGGCAGUGCACUCUGCUGACAGCAGCAAACACUGAAAGGGAGGAAUUAUCAAGC -------...............-----...((--(((..(((.((((.......))))((((....((((.....))))....)))))))..)))))........... ( -24.70, z-score = -0.98, R) >consensus ________C_CCCCU_CCCCCCACACCCCACCGAUCCACUUCCGCCCCUCGUGAAGGCAGUGCACUCUGCUGACAGCAGCAAACACAGAAAGGGAGGAAUUAUCAAGC ......................................((((((((((....)).))).(((.....(((((....)))))..))).....)))))............ (-18.34 = -18.67 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:38:27 2011