
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,377,560 – 19,377,659 |
| Length | 99 |
| Max. P | 0.700364 |

| Location | 19,377,560 – 19,377,659 |
|---|---|
| Length | 99 |
| Sequences | 11 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 83.84 |
| Shannon entropy | 0.32923 |
| G+C content | 0.50603 |
| Mean single sequence MFE | -31.00 |
| Consensus MFE | -16.70 |
| Energy contribution | -16.40 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.700364 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 19377560 99 - 27905053 -CAGGAGCAAAACGGAAGUUCGACGCCCAAGUGGCAGUCGCGAUAAUAGCGCCCUGCCAUGUGCGCUAAAGUAUUAGCAUAU--ACACAGGCGCACACGAAC------ -..((.((..(((....)))....))))..(((((((.(((.......)))..)))))))(((((((...((((......))--))...)))))))......------ ( -36.50, z-score = -3.53, R) >droPer1.super_7 2960380 99 + 4445127 -CAGCAGGGAAACGGAAGUUCGACGCCCAAGUGGCAGUCGCGAUAAUAGCGCCCUGCCAUGUGCGCUAAAGUAUUAGCAUAUAAACAUACACACAUGCAC-------- -..((((((.(((....))).((((((.....))).)))((.......)).))))))((((((.(((((....))))).(((....)))..))))))...-------- ( -31.30, z-score = -2.33, R) >dp4.chr2 2866787 99 - 30794189 -CAGCAGGGAAACGGAAGUUCGACGCCCAAGUGGCAGUCGCGAUAAUAGCGCCCUGCCAUGUGCGCUAAAGUAUUAGCAUAUAAACAUACACACAUGCAC-------- -..((((((.(((....))).((((((.....))).)))((.......)).))))))((((((.(((((....))))).(((....)))..))))))...-------- ( -31.30, z-score = -2.33, R) >droAna3.scaffold_13340 3529066 108 + 23697760 CUGCUGGCAAAACGGAAGUUCGACGCCCAAGUGGCAGUCGCGAUAAUAGGUCCCUGCCAUGUGCGCUAAAGUAUUAGCAUAUAAACAUAGACUCACACACACACUCAC ..((.(((..(((....)))....)))...(((((((..((........))..)))))))..))(((((....))))).............................. ( -25.10, z-score = -0.09, R) >droEre2.scaffold_4820 1805082 95 + 10470090 -----AGCAAAACGGAAGUUCGACGCCCAAGUGGCAGUCGCGAUAAUAGCGCCCUGCCAUGUGCGCUAAAGUAUUAGCAUAU--ACACAGGCGCACACGAGC------ -----............(((((........(((((((.(((.......)))..)))))))(((((((...((((......))--))...))))))).)))))------ ( -34.50, z-score = -2.79, R) >droYak2.chr3R 20264022 99 - 28832112 -CAGGAGCAAAACGGAAGUUCGACGCCCAAGUGGCAGUCGCGAUAAUAGCGCCCUGCCAUGUGCGCUAAAGUAUUAGCAUAU--ACACAGGCGCACACGAAC------ -..((.((..(((....)))....))))..(((((((.(((.......)))..)))))))(((((((...((((......))--))...)))))))......------ ( -36.50, z-score = -3.53, R) >droSec1.super_0 19738363 99 - 21120651 -CAGGAGCAAAACGGAAGUUCGACGCCCAAGUGGCAGUCGCGAUAAUAGCGCCCUGCCAUGUGCGCUAAAGUAUUAGCAUAU--ACACAGGCGCACACGAAC------ -..((.((..(((....)))....))))..(((((((.(((.......)))..)))))))(((((((...((((......))--))...)))))))......------ ( -36.50, z-score = -3.53, R) >droSim1.chr3R 19200315 99 - 27517382 -CAGGAGCAAAACGGAAGUUCGACGCCCAAGUGGCAGUCGCGAUAAUAGCGCCCUGCCAUGUGCGCUAAAGUAUUAGCAUAU--ACACAGGCGCACACGAAC------ -..((.((..(((....)))....))))..(((((((.(((.......)))..)))))))(((((((...((((......))--))...)))))))......------ ( -36.50, z-score = -3.53, R) >droGri2.scaffold_14906 9921051 94 + 14172833 -CAGCAGGAAAACGGAAGUUCGACGCCCAAGUGGCAGCUGCGAUAAUAGUGCUCAG-CAUGUGUGCUAAAGUAUUAGCAUAUAAACAUACACACAC------------ -..((((...(((....)))....(((.....)))..)))).......(((.....-..((((((((((....))))))))))......)))....------------ ( -24.42, z-score = -1.11, R) >droMoj3.scaffold_6540 2494209 98 + 34148556 -CAGCAGGAAAGCGGAAGUUCGACGCCUAAGUGGCAGACCCGAUAAUAGUGCUCGG-CAUGUGUGCUAAAGUAUUAGCAUAUAAACAUAUAAACACACAC-------- -..((.....(((....)))(((.(((((..(((.....)))....))).))))))-).((((((((((....)))))))))).................-------- ( -22.20, z-score = -0.62, R) >droVir3.scaffold_13047 8999702 104 - 19223366 -CAGCAGGAUGGCGGAAGUUCGACGCCCAAGUGGCACCCGCGAUAAUAGUGCUCGG-CAUGUGUGCUAAAGUAUUAGCAUAUAAACAUAUAAACACGAACACACAC-- -..((......))....(((((..(((.....(((((...........))))).))-).((((((((((....))))))))))............)))))......-- ( -26.20, z-score = -0.58, R) >consensus _CAGCAGCAAAACGGAAGUUCGACGCCCAAGUGGCAGUCGCGAUAAUAGCGCCCUGCCAUGUGCGCUAAAGUAUUAGCAUAUAAACAUAGACACACACAAAC______ ..........(((....)))....(((.....)))....((((((.((((((.(......).))))))...)))).)).............................. (-16.70 = -16.40 + -0.30)

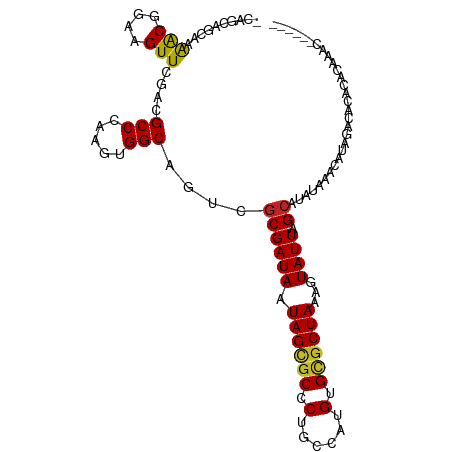
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:38:25 2011