
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 19,335,439 – 19,335,545 |
| Length | 106 |
| Max. P | 0.723485 |

| Location | 19,335,439 – 19,335,545 |
|---|---|
| Length | 106 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 71.74 |
| Shannon entropy | 0.59382 |
| G+C content | 0.51064 |
| Mean single sequence MFE | -31.76 |
| Consensus MFE | -15.40 |
| Energy contribution | -14.13 |
| Covariance contribution | -1.27 |
| Combinations/Pair | 1.63 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.723485 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
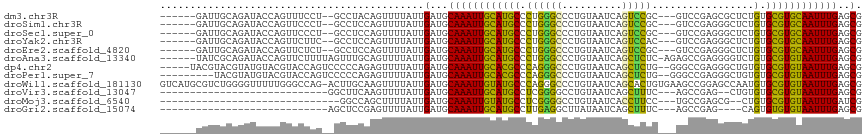
>dm3.chr3R 19335439 106 - 27905053 ------GAUUGCAGAUACCAGUUUCCU--GCCUACAGUUUUAUUGAUGCAAAUUGCAUGCCCUGGGCCCUGUAAUCAGUCCGC---GUCCGAGCGCUCUGUGCGUGCAAUUUGAGCG ------....((......((((...((--(....)))....))))...((((((((((((.(.((((.(((....)))...((---......)))))).).)))))))))))).)). ( -31.70, z-score = -1.37, R) >droSim1.chr3R 19154976 106 - 27517382 ------GAUUGCAGAUACCAGUUCCCU--GCCUCCAGUUUUAUUGAUGCAAAUUGCAUGCCCUGGGCCCUGUAAUCAGUCCGC---GUCCGAGGGCUCUGUGCGUGCAAUUUGAGCG ------....((......((((...((--(....)))....))))...((((((((((((.(.(((((((.............---.....))))))).).)))))))))))).)). ( -33.87, z-score = -1.60, R) >droSec1.super_0 19698284 106 - 21120651 ------GAUUGCAGAUACCAGUUCCCU--GCCUCCAGUUUUAUUGAUGCAAAUUGCAUGCCCUGGGCCCUGUAAUCAGUCCGC---GUCCGAGGGCUCUGUGCGUGCAAUUUGAGCG ------....((......((((...((--(....)))....))))...((((((((((((.(.(((((((.............---.....))))))).).)))))))))))).)). ( -33.87, z-score = -1.60, R) >droYak2.chr3R 20220304 106 - 28832112 ------GAUUGCAGAUACCAGUUCUUC--GCCUCCAGUUUUAUUGAUGCAAAUUGCAUGCCCUGGGCCCUGUAAUCAGUCCAC---GUCCGAGGGCUCUGUGCGUGCAAUUUGAGCG ------....((......((((....(--.......)....))))...((((((((((((.(.(((((((.............---.....))))))).).)))))))))))).)). ( -31.97, z-score = -1.21, R) >droEre2.scaffold_4820 1758275 106 + 10470090 ------GAUUGCAGAUACCAGUUCUCU--GCCUCCAGUUUUAUUGAUGCAAAUUGCAUGCCCUGGGCCCUGUAAUCAGUCCGC---GUCCGAGGGCUCUGUGCGUGCAAUUUGAGCG ------....((......((((...((--(....)))....))))...((((((((((((.(.(((((((.............---.....))))))).).)))))))))))).)). ( -34.57, z-score = -1.65, R) >droAna3.scaffold_13340 3482302 110 + 23697760 ------UAUCGCAGAUACCAGUUCUUUUAGUUUGCAGUUUUAUUGAUGCAAAUUGCAUGCCCUGGGCCCUGUAAUCAGCUCUC-AGAGCCGAGGGGUCUGUGCGUGUAAUUUGAGCG ------....((((((.............)))))).........(...((((((((((((.(.(((((((....((.((((..-.)))).)))))))))).))))))))))))..). ( -35.32, z-score = -1.80, R) >dp4.chr2 2824234 110 - 30794189 -----UACGUACGUAUGUACGUACCAGUCCCCCAGAGUUUUAUUGAUGCAAAUUGCACGCCCAGGGCCCUGUAAUCAGCUCUG--GGGCCGAGGGCUGUGUGCGUGUAAUUUGAGCG -----(((((((....))))))).....................(...((((((((((((.((.(((((.....((.(((...--.))).))))))).)).))))))))))))..). ( -41.40, z-score = -2.28, R) >droPer1.super_7 2917676 106 + 4445127 ---------UACGUAUGUACGUACCAGUCCCCCAGAGUUUUAUUGAUGCAAAUUGCACGCCCAGGGCCCUGUAAUCAGCUCUG--GGGCCGAGGGCUGUGUGCGUGUAAUUUGAGCG ---------(((((....))))).....................(...((((((((((((.((.(((((.....((.(((...--.))).))))))).)).))))))))))))..). ( -37.30, z-score = -1.50, R) >droWil1.scaffold_181130 7269808 116 + 16660200 GUCAUGCGUCUGGGGUUUUUGGGCCAG-ACUUGCAAGUUUUAUUGAUGCAAAUUGUAUGCCCAGGGCCCUGUAAUCAGCACUGUGAAGCCGGAGCCAAUGUGCGUGUAAUUUGAGCG .....((((((((..........))))-))..................((((((((((((.((.(((((.((..(((......))).)).)).)))..)).)))))))))))).)). ( -31.10, z-score = 1.34, R) >droVir3.scaffold_13047 8964386 84 - 19223366 ----------------------------GGCUUCAAGUUUUAUUGAUGCAAAUUGCAUGCCUCGGGGCCUGUAAUCAGCUUUC---AGCCGAG--CUGUGUGCGUGUAAUUUGAGCG ----------------------------.(((((((......))))..(((((((((((((....)))..(((..((((((..---....)))--)))..)))))))))))))))). ( -27.00, z-score = -1.19, R) >droMoj3.scaffold_6540 2452354 82 + 34148556 ------------------------------GGCCAGCUUUUAUUGAUGCAAAUUGUAUGCCUCGGGGCCUGUAAUCACCUUCC---UGCCGAGCG--CUGUGCGUGUAAUUUGAUCG ------------------------------..............(((.((((((((((((..(((.(((.(((..........---))).).)).--))).))))))))))))))). ( -20.60, z-score = 0.17, R) >droGri2.scaffold_15074 7073206 82 + 7742996 ----------------------------AGCUCCGAGUUUUAUUGAUGCAAAUUGCAUGCCUUGAGGCUUAUAAUCAGCUUUC---AGCCGAG----CAGUGUGUGUAAUUUGAGCG ----------------------------.(((((((......)))....(((((((((((.(((.((((..............---))))...----))).))))))))))))))). ( -22.44, z-score = -0.99, R) >consensus ______GAUUGCAGAUACCAGUUCCCU__GCCUCCAGUUUUAUUGAUGCAAAUUGCAUGCCCUGGGCCCUGUAAUCAGCUCUC___GGCCGAGGGCUCUGUGCGUGUAAUUUGAGCG ............................................(...((((((((((((.((((((..........))))).................).))))))))))))..). (-15.40 = -14.13 + -1.27)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:38:21 2011